1YU6
 
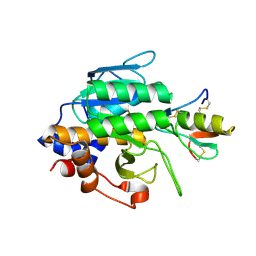 | | Crystal Structure of the Subtilisin Carlsberg:OMTKY3 Complex | | Descriptor: | CALCIUM ION, Ovomucoid, Subtilisin Carlsberg | | Authors: | Maynes, J.T, Cherney, M.M, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2005-02-11 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the subtilisin Carlsberg-OMTKY3 complex reveals two different ovomucoid conformations.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3SY4
 
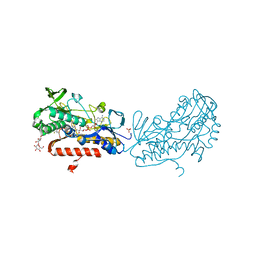 | | Crystal structure of sulfide:quinone oxidoreductase Ser126Ala variant from Acidithiobacillus ferrooxidans | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-15 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3SYI
 
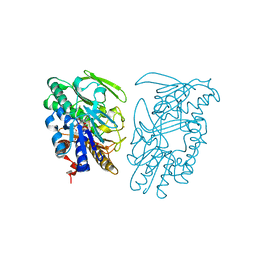 | | Crystal structure of sulfide:quinone oxidoreductase Ser126Ala variant from Acidithiobacillus ferrooxidans using 7.0 keV diffraction data | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2001 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3SZF
 
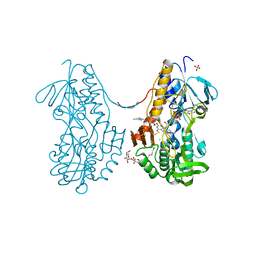 | | Crystal structure of sulfide:quinone oxidoreductase H198A variant from Acidithiobacillus ferrooxidans in complex with bound trisulfide and decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-18 | | Release date: | 2012-05-16 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2.0994 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3SZW
 
 | | Crystal structure of sulfide:quinone oxidoreductase Cys128Ser variant from Acidithiobacillus ferrooxidans in complex with decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-19 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3SX6
 
 | | Crystal structure of sulfide:quinone oxidoreductase Cys356Ala variant from Acidithiobacillus ferrooxidans complexed with decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-14 | | Release date: | 2012-05-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7955 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3SXI
 
 | | Crystal structure of sulfide:quinone oxidoreductase Cys128Ala variant from Acidithiobacillus ferrooxidans complexed with decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-14 | | Release date: | 2012-05-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1792 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3T2K
 
 | | Crystal structure of sulfide:quinone oxidoreductase Cys128Ala variant from Acidithiobacillus ferrooxidans with bound trisulfane | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-22 | | Release date: | 2012-05-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3T0K
 
 | | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans with bound trisulfide and decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-20 | | Release date: | 2012-05-16 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3SZ0
 
 | | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans in complex with sodium selenide | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, SELENIUM ATOM, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-18 | | Release date: | 2012-05-16 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.1501 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3SZC
 
 | | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans in complex with gold (I) cyanide | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-18 | | Release date: | 2012-05-16 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3T2Y
 
 | | Crystal structure of sulfide:quinone oxidoreductase His132Ala variant from Acidithiobacillus ferrooxidans with bound disulfide | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-23 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (2.5001 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
2ATO
 
 | | Crystal structure of Human Cathepsin K in complex with myocrisin | | Descriptor: | (S)-(1,2-DICARBOXYETHYLTHIO)GOLD, Cathepsin K, SULFATE ION | | Authors: | Weidauer, E, Yasuda, Y, Biswal, B.K, Kerr, L.D, Cherney, M.M, Gordon, R.E, James, M.N.G, Bromme, D. | | Deposit date: | 2005-08-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of disease-modifying anti-rheumatic drugs (DMARDs) on the activities of rheumatoid arthritis-associated cathepsins K and S.
Biol.Chem., 388, 2007
|
|
3T14
 
 | | Crystal structure of sulfide:quinone oxidoreductase Cys128Ala variant from Acidithiobacillus ferrooxidans with bound disulfide | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-21 | | Release date: | 2012-05-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3EI9
 
 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with L-Glu: External aldimine form | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-glutamic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EI8
 
 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with LL-DAP: External aldimine form | | Descriptor: | (2S,6S)-2-amino-6-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}heptanedioic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EIB
 
 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana | | Descriptor: | GLYCEROL, LL-diaminopimelate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EI6
 
 | | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with PLP-DAP: an external aldimine mimic | | Descriptor: | (2S,6S)-2-amino-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]heptanedioic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EJX
 
 | | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana in complex with LL-AziDAP | | Descriptor: | (2S,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, Diaminopimelate epimerase, chloroplastic | | Authors: | Pillai, B, Moorthie, V.A, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana, an amino acid racemase critical for L-lysine biosynthesis.
J.Mol.Biol., 385, 2009
|
|
3EKM
 
 | | Crystal structure of diaminopimelate epimerase form arabidopsis thaliana in complex with irreversible inhibitor DL-AziDAP | | Descriptor: | (2R,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, Diaminopimelate epimerase, chloroplastic | | Authors: | Pillai, B, Moorthie, V.A, Cherney, M.M, van Belkum, M.J, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-19 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana, an amino acid racemase critical for L-lysine biosynthesis.
J.Mol.Biol., 385, 2009
|
|
3EI5
 
 | | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with PLP-Glu: an external aldimine mimic | | Descriptor: | GLYCEROL, LL-diaminopimelate aminotransferase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
2Z7A
 
 | | X-ray crystal structure of RV0760c from Mycobacterium tuberculosis at 2.10 Angstrom resolution | | Descriptor: | ACETATE ION, Putative steroid isomerase | | Authors: | Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-08-17 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase.
Biochim.Biophys.Acta, 1784, 2008
|
|
2Z20
 
 | | Crystal structure of LL-Diaminopimelate Aminotransferase from Arabidopsis thaliana | | Descriptor: | GLYCEROL, LL-diaminopimelate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Watanabe, N, Cherney, M.M, van Belkum, M.J, Marcus, S.L, Flegel, M.D, Clay, M.D, Deyholos, M.K, Vederas, J.C, James, M.N.G. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana: a recently discovered enzyme in the biosynthesis of L-lysine by plants and Chlamydia
J.Mol.Biol., 371, 2007
|
|
2Z77
 
 | | X-ray crystal structure of RV0760c from Mycobacterium tuberculosis in complex with estradiol-17beta-hemisuccinate | | Descriptor: | 4-{[(14beta,17alpha)-3-hydroxyestra-1,3,5(10)-trien-17-yl]oxy}-4-oxobutanoic acid, ACETATE ION, NICOTINAMIDE, ... | | Authors: | Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-08-16 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase.
Biochim.Biophys.Acta, 1784, 2008
|
|
3EIA
 
 | | Crystal structure of K270Q variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with L-Glu: External aldimine form | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-glutamic acid, LL-diaminopimelate aminotransferase, SULFATE ION | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
