3LMY
 
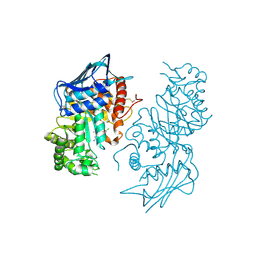 | | The Crystal Structure of beta-hexosaminidase B in complex with Pyrimethamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bateman, K.S, Cherney, M.M, Withers, S.G, Mahuran, D.J, Tropak, M, James, M.N.G. | | Deposit date: | 2010-02-01 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of beta-hexosaminidase B in complex with Pyrimethamine
To be Published
|
|
2ALP
 
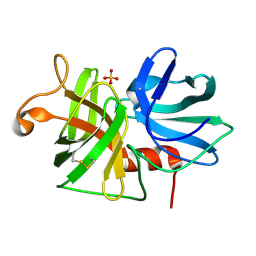 | | REFINED STRUCTURE OF ALPHA-LYTIC PROTEASE AT 1.7 ANGSTROMS RESOLUTION. ANALYSIS OF HYDROGEN BONDING AND SOLVENT STRUCTURE | | Descriptor: | ALPHA-LYTIC PROTEASE, SULFATE ION | | Authors: | Fujinaga, M, Delbaere, L.T.J, Brayer, G.D, James, M.N.G. | | Deposit date: | 1985-03-07 | | Release date: | 1985-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined structure of alpha-lytic protease at 1.7 A resolution. Analysis of hydrogen bonding and solvent structure.
J.Mol.Biol., 184, 1985
|
|
4OBS
 
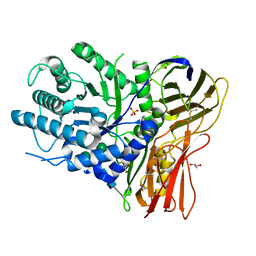 | | Crystal structure of human alpha-L-iduronidase in the P212121 form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, CHLORIDE ION, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, James, M.N.G. | | Deposit date: | 2014-01-07 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of human alpha-L-iduronidase in the P212121 form
To be Published
|
|
4OBR
 
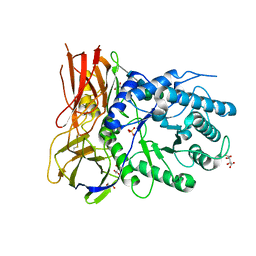 | | Crystal structure of human alpha-L-iduronidase complex with alpha-L-iduronic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, ... | | Authors: | Bie, H.Y, Yin, J, He, X, Kermode, A.R, James, M.N.G. | | Deposit date: | 2014-01-07 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of human alpha-L-iduronidase complex with alpha-L-iduronic acid
To be Published
|
|
4Q0G
 
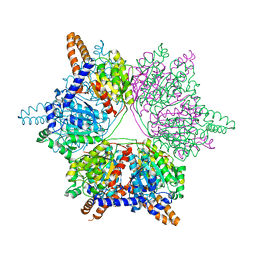 | |
2WEA
 
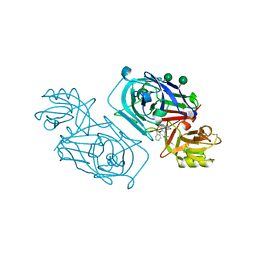 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE INHIBITOR: METHYL[CYCLO-7[(2R)-((N-VALYL) AMINO)-2-(HYDROXYL-(1S)-1-METHYOXYCARBONYL-2-PHENYLETHOXY) PHOSPHINYLOXY-ETHYL]-1-NAPHTHALENEACETAMIDE], SODIUM SALT | | Descriptor: | METHYL[CYCLO-7[(2R)-((N-VALYL)AMINO)-2-(HYDROXYL-(1S)-1-METHYLOXYCARBONYL-2-PHENYLETHOXY)PHOSPHINYLOXY-ETHYL]-1-NAPHTHALENEACETAMIDE], PENICILLOPEPSIN, SULFATE ION, ... | | Authors: | Ding, J, Fraser, M.E, James, M.N.G. | | Deposit date: | 1998-02-03 | | Release date: | 1998-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Macrocyclic Inhibitors of Penicillopepsin. II. X-Ray Crystallographic Analyses of Penicillopepsin Complexed with a P3-P1 Macrocyclic Peptidyl Inhibitor and with its Two Acyclic Analogues
J.Am.Chem.Soc., 120, 1998
|
|
2WED
 
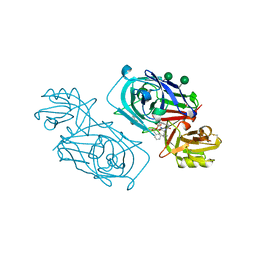 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE MACROCYCLIC INHIBITOR:METHYL[CYCLO-7[(2R)-((N-VALYL)AMINO)-2-(HYDROXYL-(1S)-1-METHYOXYCARBONYL-2-PHENYLETHOXY)PHOSPHINYLOXY-ETHYL]-1-NAPHTHALENEACETAMIDE], SODIUM SALT | | Descriptor: | METHYL[CYCLO-7[(2R)-((N-VALYL)AMINO)-2-(HYDROXYL-(1S)-1-METHYLOXYCARBONYL-2-PHENYLETHOXY)PHOSPHINYLOXY-ETHYL]-1-NAPHTHALENEACETAMIDE], PENICILLOPEPSIN, SULFATE ION, ... | | Authors: | Ding, J, Fraser, M.E, James, M.N.G. | | Deposit date: | 1998-02-03 | | Release date: | 1998-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Macrocyclic Inhibitors of Penicillopepsin. II. X-Ray Crystallographic Analyses of Penicillopepsin Complexed with a P3-P1 Macrocyclic Peptidyl Inhibitor and with its Two Acyclic Analogues
J.Am.Chem.Soc., 120, 1998
|
|
3AAY
 
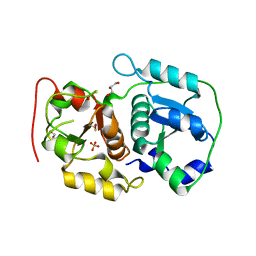 | | Crystal structure of probable thiosulfate sulfurtransferase CYSA3 (RV3117) from Mycobacterium tuberculosis: orthorhombic form | | Descriptor: | GLYCEROL, Putative thiosulfate sulfurtransferase, SULFATE ION | | Authors: | Sankaranarayanan, R, Witholt, S.J, Cherney, M.M, Garen, C.R, Cherney, L.T, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of probable thiosulfate sulfurtransferase CysA3 (Rv3117) from Mycobacterium tuberculosis
To be Published
|
|
3AAX
 
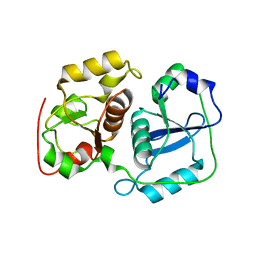 | | Crystal structure of probable thiosulfate sulfurtransferase cysa3 (RV3117) from Mycobacterium tuberculosis: monoclinic FORM | | Descriptor: | Putative thiosulfate sulfurtransferase | | Authors: | Sankaranarayanan, R, Witholt, S.J, Cherney, M.M, Garen, C.R, Cherney, L.T, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of probable thiosulfate sulfurtransferase CysA3 (Rv3117) from Mycobacterium tuberculosis
To be Published
|
|
3NAG
 
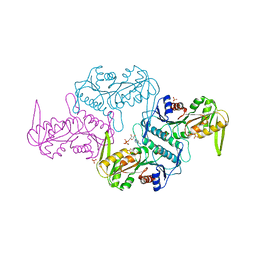 | | Crystal structure of the phosphoribosylpyrophosphate (PRPP) synthetase from Thermoplasma Volcanium in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribose-phosphate pyrophosphokinase, ... | | Authors: | Cherney, M.M, Cherney, L.T, Garen, C.R, James, M.N.G. | | Deposit date: | 2010-06-02 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of Thermoplasma volcanium phosphoribosyl pyrophosphate synthetase bound to ribose-5-phosphate and ATP analogs.
J.Mol.Biol., 413, 2011
|
|
1PPK
 
 | |
1BXO
 
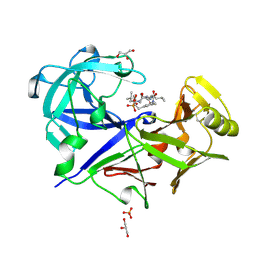 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE INHIBITOR: METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUT YL] HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL) PHENYLPROPANOATE | | Descriptor: | GLYCEROL, METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUTYL]HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL)PHENYLPROPANOATE, PROTEIN (PENICILLOPEPSIN), ... | | Authors: | Khan, A.R, Parrish, J.C, Fraser, M.E, Smith, W.W, Bartlett, P.A, James, M.N.G. | | Deposit date: | 1998-10-07 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes.
Biochemistry, 37, 1998
|
|
1BXQ
 
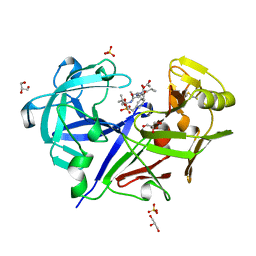 | | ACID PROTEINASE (PENICILLOPEPSIN) COMPLEX WITH PHOSPHONATE INHIBITOR. | | Descriptor: | 2-[(1R)-1-(N-(3-METHYLBUTANOYL)-L-VALYL-L-ASPARAGINYL)-AMINO)-3-METHYLBUTYL]HYDROXYPHOSPHINYLOXY]-3-PHENYLPROPANOIC ACID METHYLESTER, ACETATE ION, GLYCEROL, ... | | Authors: | Parrish, J.C, Khan, A.R, Fraser, M.E, Smith, W.W, Bartlett, P.A, James, M.N.G. | | Deposit date: | 1998-10-07 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes.
Biochemistry, 37, 1998
|
|
4J8R
 
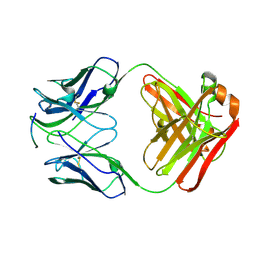 | | Structure of an octapeptide repeat of the prion protein bound to the POM2 Fab antibody fragment | | Descriptor: | Heavy chain of POM2 Fab, Light chain of POM2 Fab, Major prion protein | | Authors: | Swayampakula, M, Baral, P.K, Kav, N.N.V, Aguzzi, A, James, M.N.G. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | The crystal structure of an octapeptide repeat of the Prion protein in complex with a Fab fragment of the POM2 antibody.
Protein Sci., 22, 2013
|
|
4KGJ
 
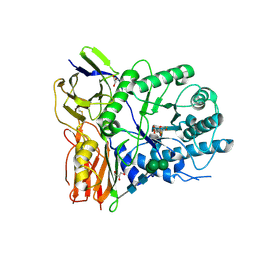 | | Crystal structure of human alpha-L-iduronidase complex with 5-fluoro-alpha-L-idopyranosyluronic acid fluoride | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-fluoro-alpha-L-idopyranosyluronic acid fluoride, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
4KH2
 
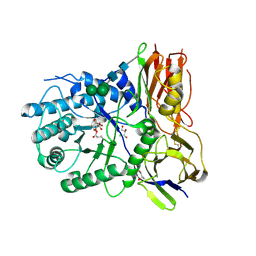 | | Crystal structure of human alpha-L-iduronidase complex with 2-deoxy-2-fluoro-alpha-L-idopyranosyluronic acid fluoride | | Descriptor: | 2,6-anhydro-5-deoxy-5-fluoro-L-idonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
4KGL
 
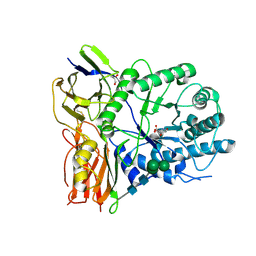 | | Crystal structure of human alpha-L-iduronidase complex with [2R,3R,4R,5S]-2-carboxy-3,4,5-trihydroxy-piperidine | | Descriptor: | (2R,3R,4R,5S)-3,4,5-trihydroxypiperidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
2SCU
 
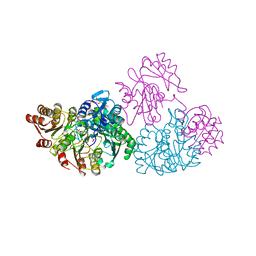 | | A detailed description of the structure of Succinyl-COA synthetase from Escherichia coli | | Descriptor: | COENZYME A, PROTEIN (SUCCINYL-COA LIGASE), SULFATE ION | | Authors: | Fraser, M.E, Wolodko, W.T, James, M.N.G, Bridger, W.A. | | Deposit date: | 1998-09-24 | | Release date: | 1999-08-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A detailed structural description of Escherichia coli succinyl-CoA synthetase.
J.Mol.Biol., 285, 1999
|
|
2GT7
 
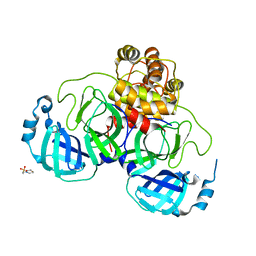 | | Crystal structure of SARS coronavirus main peptidase at pH 6.0 in the space group P21 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C-like proteinase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N.G. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase.
J.Mol.Biol., 366, 2007
|
|
2GT8
 
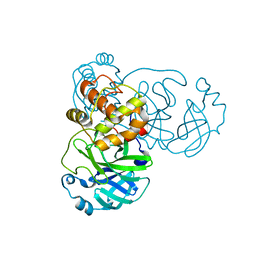 | | Crystal structure of SARS coronavirus main peptidase (with an additional Ala at the N-terminus of each protomer) in the space group P43212 | | Descriptor: | 3C-like proteinase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N.G. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase.
J.Mol.Biol., 366, 2007
|
|
2SGQ
 
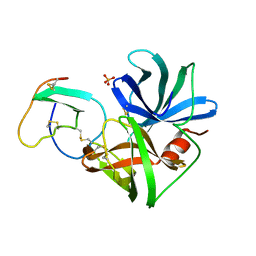 | | GLN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
2SGF
 
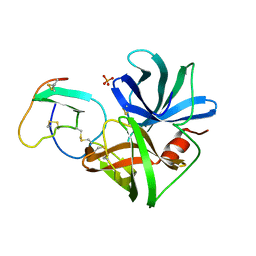 | | PHE 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
2SGD
 
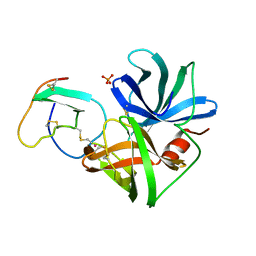 | | ASP 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 | | Descriptor: | Ovomucoid, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
4YX2
 
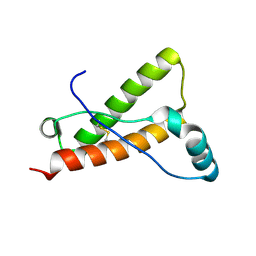 | | Crystal structure of Bovine prion protein complexed with POM1 FAB | | Descriptor: | Major prion protein, POM1 FAB HEAVY CHAIN, POM1 FAB LIGHT CHAIN | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2015-03-22 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | X-ray structural and molecular dynamical studies of the globular domains of cow, deer, elk and Syrian hamster prion proteins.
J.Struct.Biol., 192, 2015
|
|
4YXL
 
 | | Crystal structure of Syrian hamster prion protein complexed with POM1 FAB | | Descriptor: | Major prion protein, POM1 FAB HEAVY CHAIN, POM1 FAB LIGHT CHAIN, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | X-ray structural and molecular dynamical studies of the globular domains of cow, deer, elk and Syrian hamster prion proteins.
J.Struct.Biol., 192, 2015
|
|
