2PSG
 
 | |
2SGA
 
 | |
2A5A
 
 | | Crystal structure of unbound SARS coronavirus main peptidase in the space group C2 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like peptidase, CHLORIDE ION | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2A5I
 
 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in the space group C2 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 1,2-ETHANEDIOL, 3C-like peptidase, ... | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2A5K
 
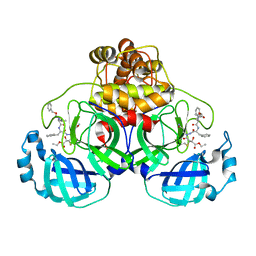 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in space group P212121 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 3C-like peptidase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2AQ6
 
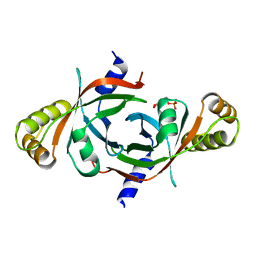 | | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase complexed with pyridoxal 5'-phosphate at 1.7 a resolution | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRIDOXINE 5'-PHOSPHATE OXIDASE | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Mycobacterium tuberculosispyridoxine 5'-phosphate oxidase and its complexes with flavin mononucleotide and pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4DGI
 
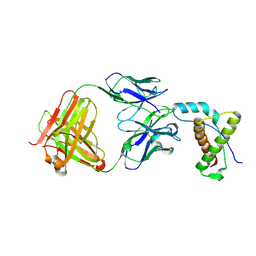 | | Structure of POM1 FAB fragment complexed with human PrPc Fragment 120-230 | | Descriptor: | Major prion protein, POM1 Fab Heavy chain, POM1 Fab Light chain, ... | | Authors: | Baral, P.K, Wieland, B, Swayampakula, M, James, M.N. | | Deposit date: | 2012-01-26 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies on the folded domain of the human prion protein bound to the Fab fragment of the antibody POM1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3IT4
 
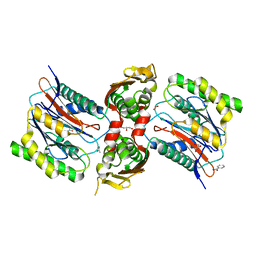 | | The Crystal Structure of Ornithine Acetyltransferase from Mycobacterium tuberculosis (Rv1653) at 1.7 A | | Descriptor: | ACETATE ION, Arginine biosynthesis bifunctional protein argJ alpha chain, Arginine biosynthesis bifunctional protein argJ beta chain, ... | | Authors: | Sankaranarayanan, R, Cherney, M.M, Garen, C, Garen, G, Yuan, M, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-08-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular structure of ornithine acetyltransferase from Mycobacterium tuberculosis bound to ornithine, a competitive inhibitor.
J.Mol.Biol., 397, 2010
|
|
3IT6
 
 | | The Crystal Structure of Ornithine Acetyltransferase complexed with Ornithine from Mycobacterium tuberculosis (Rv1653) at 2.4 A | | Descriptor: | Arginine biosynthesis bifunctional protein argJ alpha chain, Arginine biosynthesis bifunctional protein argJ beta chain, L-ornithine | | Authors: | Sankaranarayanan, R, Cherney, M.M, Garen, C, Garen, G, Yuan, M, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-08-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular structure of ornithine acetyltransferase from Mycobacterium tuberculosis bound to ornithine, a competitive inhibitor.
J.Mol.Biol., 397, 2010
|
|
4H88
 
 | | Structure of POM1 FAB fragment complexed with mouse PrPc Fragment 120-230 | | Descriptor: | Major prion protein, POM1 FAB CHAIN H, POM1 FAB CHAIN L, ... | | Authors: | Baral, P.K, Wieland, B, Swayampakula, M, James, M.N. | | Deposit date: | 2012-09-21 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The toxicity of antiprion antibodies is mediated by the flexible tail of the prion protein.
Nature, 501, 2013
|
|
2HAL
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | Hepatitis A Protease 3C, N-ACETYL-LEUCYL-PHENYLALANYL-PHENYLALANYL-GLUTAMATE-FLUOROMETHYLKETONE INHIBITOR, N-[(BENZYLOXY)CARBONYL]-L-ALANINE | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
J.Mol.Biol., 361, 2006
|
|
1Y30
 
 | | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase complexed with flavin mononucleotide at 2.2 a resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein Rv1155 | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-23 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Mycobacterium tuberculosispyridoxine 5'-phosphate oxidase and its complexes with flavin mononucleotide and pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Y0H
 
 | | Structure of Rv0793 from Mycobacterium tuberculosis | | Descriptor: | ACETATE ION, hypothetical protein Rv0793 | | Authors: | Lemieux, M.J, Ference, C, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-15 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Rv0793, a hypothetical monooxygenase from M. tuberculosis
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
3T31
 
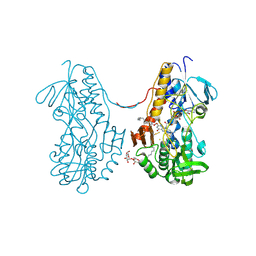 | | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans in complex with decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cherney, M.M, Zhang, Y, Solomonson, M, Weiner, J.H, James, M.N. | | Deposit date: | 2011-07-24 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans: insights into sulfidotrophic respiration and detoxification.
J.Mol.Biol., 398, 2010
|
|
3T2Z
 
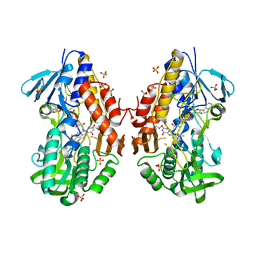 | | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans | | Descriptor: | 1,3-BUTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, Solomonson, M, Weiner, J.H, James, M.N. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2994 Å) | | Cite: | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans: insights into sulfidotrophic respiration and detoxification.
J.Mol.Biol., 398, 2010
|
|
1JAK
 
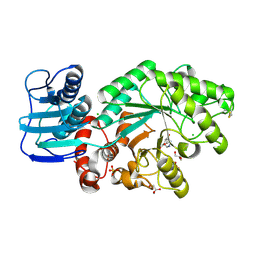 | | Streptomyces plicatus beta-N-acetylhexosaminidase in Complex with (2R,3R,4S,5R)-2-acetamido-3,4-dihydroxy-5-hydroxymethyl-piperidinium chloride (IFG) | | Descriptor: | (2R,3R,4S,5R)-2-ACETAMIDO-3,4-DIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Mark, B.L, Vocadlo, D.J, Zhao, D, Knapp, S, Withers, S.G, James, M.N. | | Deposit date: | 2001-05-30 | | Release date: | 2001-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural assessment of the 1-N-azasugar GalNAc-isofagomine as a potent family 20 beta-N-acetylhexosaminidase inhibitor.
J.Biol.Chem., 276, 2001
|
|
2BDX
 
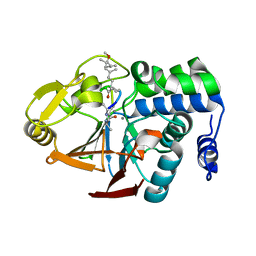 | | X-ray Crystal Structure of dihydromicrocystin-LA bound to Protein Phosphatase-1 | | Descriptor: | DIHYDROMICROCYSTIN-LA, MANGANESE (II) ION, Serine/threonine protein phosphatase PP1-gamma catalytic subunit | | Authors: | Maynes, J.T, Luu, H.A, Cherney, M.M, Andersen, R.J, Williams, D, Holmes, C.F, James, M.N. | | Deposit date: | 2005-10-21 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Protein Phosphatase-1 Bound to Motuporin and Dihydromicrocystin-LA: Elucidation of the Mechanism of Enzyme Inhibition by Cyanobacterial Toxins.
J.Mol.Biol., 356, 2006
|
|
2BCD
 
 | | X-ray crystal structure of Protein Phosphatase-1 with the marine toxin motuporin bound | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, MOTUPORIN, ... | | Authors: | Maynes, J.T, Luu, H.A, Cherney, M.M, Andersen, R.J, Williams, D, Holmes, C.F, James, M.N. | | Deposit date: | 2005-10-19 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Protein Phosphatase-1 Bound to Motuporin and Dihydromicrocystin-LA: Elucidation of the Mechanism of Enzyme Inhibition by Cyanobacterial Toxins.
J.Mol.Biol., 356, 2006
|
|
2ASF
 
 | | Crystal structure of the conserved hypothetical protein Rv2074 from Mycobacterium tuberculosis 1.6 A | | Descriptor: | CITRIC ACID, Hypothetical protein Rv2074, SODIUM ION | | Authors: | Biswal, B.K, Au, K, Cherney, M.M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The molecular structure of Rv2074, a probable pyridoxine 5'-phosphate oxidase from Mycobacterium tuberculosis, at 1.6 angstroms resolution.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2H6M
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | N-ACETYL-LEUCYL-ALANYL-ALANYL-(N,N-DIMETHYL)-GLUTAMINE-(1,4-DIOXO-3,4-DIHYDRO-1H-PHTHALAZIN-2-YL)METHYLKETONE INHIBITOR, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Picornain 3C | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-05-31 | | Release date: | 2006-08-08 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors.
J.Mol.Biol., 361, 2006
|
|
3ASB
 
 | |
3ASA
 
 | |
2H9H
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | Hepatitis A virus protease 3C, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Three residue peptide | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-06-09 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
J.Mol.Biol., 361, 2006
|
|
1DM0
 
 | | SHIGA TOXIN | | Descriptor: | SHIGA TOXIN A SUBUNIT, SHIGA TOXIN B SUBUNIT | | Authors: | Fraser, M.E, Chernaia, M.M, Kozlov, Y.V, James, M.N. | | Deposit date: | 1999-12-13 | | Release date: | 1999-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the holotoxin from Shigella dysenteriae at 2.5 A resolution.
Nat.Struct.Biol., 1, 1994
|
|
3ERE
 
 | | Crystal structure of the arginine repressor protein from Mycobacterium tuberculosis in complex with the DNA operator | | Descriptor: | 5'-D(*DTP*DTP*DGP*DCP*DAP*DTP*DAP*DAP*DCP*DGP*DAP*DTP*DGP*DCP*DAP*DA)-3', 5'-D(*DTP*DTP*DGP*DCP*DAP*DTP*DCP*DGP*DTP*DTP*DAP*DTP*DGP*DCP*DAP*DA)-3', Arginine repressor, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Lu, G.J, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the arginine repressor protein in complex with the DNA operator from Mycobacterium tuberculosis.
J.Mol.Biol., 384, 2008
|
|
