3SZ0
 
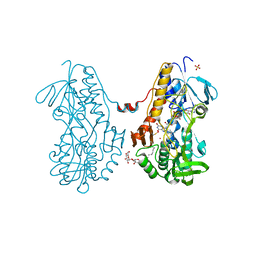 | | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans in complex with sodium selenide | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, SELENIUM ATOM, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-18 | | Release date: | 2012-05-16 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.1501 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3SX6
 
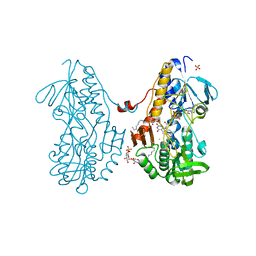 | | Crystal structure of sulfide:quinone oxidoreductase Cys356Ala variant from Acidithiobacillus ferrooxidans complexed with decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-14 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7955 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
2Z7A
 
 | | X-ray crystal structure of RV0760c from Mycobacterium tuberculosis at 2.10 Angstrom resolution | | Descriptor: | ACETATE ION, Putative steroid isomerase | | Authors: | Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-08-17 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase.
Biochim.Biophys.Acta, 1784, 2008
|
|
2Z20
 
 | | Crystal structure of LL-Diaminopimelate Aminotransferase from Arabidopsis thaliana | | Descriptor: | GLYCEROL, LL-diaminopimelate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Watanabe, N, Cherney, M.M, van Belkum, M.J, Marcus, S.L, Flegel, M.D, Clay, M.D, Deyholos, M.K, Vederas, J.C, James, M.N.G. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana: a recently discovered enzyme in the biosynthesis of L-lysine by plants and Chlamydia
J.Mol.Biol., 371, 2007
|
|
2Z77
 
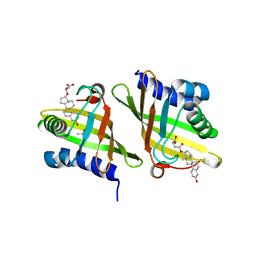 | | X-ray crystal structure of RV0760c from Mycobacterium tuberculosis in complex with estradiol-17beta-hemisuccinate | | Descriptor: | 4-{[(14beta,17alpha)-3-hydroxyestra-1,3,5(10)-trien-17-yl]oxy}-4-oxobutanoic acid, ACETATE ION, NICOTINAMIDE, ... | | Authors: | Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-08-16 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase.
Biochim.Biophys.Acta, 1784, 2008
|
|
2Z1Z
 
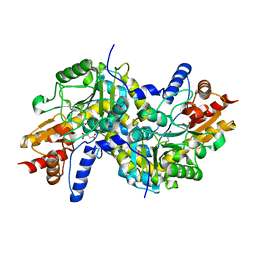 | | Crystal structure of LL-Diaminopimelate Aminotransferase from Arabidopsis thaliana complexed with L-malate ion | | Descriptor: | D-MALATE, LL-diaminopimelate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watanabe, N, Cherney, M.M, van Belkum, M.J, Marcus, S.L, Flegel, M.D, Clay, M.D, Deyholos, M.K, Vederas, J.C, James, M.N.G. | | Deposit date: | 2007-05-16 | | Release date: | 2007-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana: a recently discovered enzyme in the biosynthesis of L-lysine by plants and Chlamydia
J.Mol.Biol., 371, 2007
|
|
2SCU
 
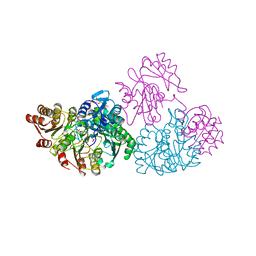 | | A detailed description of the structure of Succinyl-COA synthetase from Escherichia coli | | Descriptor: | COENZYME A, PROTEIN (SUCCINYL-COA LIGASE), SULFATE ION | | Authors: | Fraser, M.E, Wolodko, W.T, James, M.N.G, Bridger, W.A. | | Deposit date: | 1998-09-24 | | Release date: | 1999-08-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A detailed structural description of Escherichia coli succinyl-CoA synthetase.
J.Mol.Biol., 285, 1999
|
|
1PPL
 
 | |
2ZFZ
 
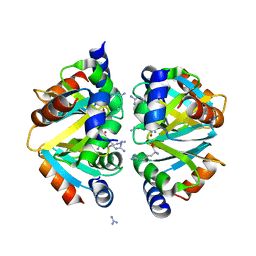 | | Crystal structure of the C-terminal domain hexamer of ArgR from Mycobacterium tuberculosis in complex with arginine | | Descriptor: | ARGININE, Arginine repressor, GUANIDINE | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Lu, G.J, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the C-terminal domain of the arginine repressor protein from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2Z76
 
 | | X-ray crystal structure of RV0760c from Mycobacterium tuberculosis at 1.82 Angstrom resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-08-16 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase.
Biochim.Biophys.Acta, 1784, 2008
|
|
3AAY
 
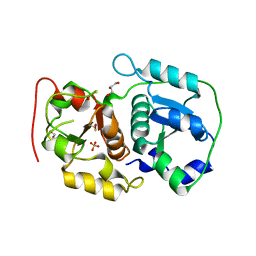 | | Crystal structure of probable thiosulfate sulfurtransferase CYSA3 (RV3117) from Mycobacterium tuberculosis: orthorhombic form | | Descriptor: | GLYCEROL, Putative thiosulfate sulfurtransferase, SULFATE ION | | Authors: | Sankaranarayanan, R, Witholt, S.J, Cherney, M.M, Garen, C.R, Cherney, L.T, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of probable thiosulfate sulfurtransferase CysA3 (Rv3117) from Mycobacterium tuberculosis
To be Published
|
|
3AAX
 
 | | Crystal structure of probable thiosulfate sulfurtransferase cysa3 (RV3117) from Mycobacterium tuberculosis: monoclinic FORM | | Descriptor: | Putative thiosulfate sulfurtransferase | | Authors: | Sankaranarayanan, R, Witholt, S.J, Cherney, M.M, Garen, C.R, Cherney, L.T, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of probable thiosulfate sulfurtransferase CysA3 (Rv3117) from Mycobacterium tuberculosis
To be Published
|
|
1PPM
 
 | |
1Y30
 
 | | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase complexed with flavin mononucleotide at 2.2 a resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein Rv1155 | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-23 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Mycobacterium tuberculosispyridoxine 5'-phosphate oxidase and its complexes with flavin mononucleotide and pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1XXO
 
 | | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase at 1.8 a resolution | | Descriptor: | hypothetical protein Rv1155 | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Garen, C, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-07 | | Release date: | 2004-11-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase at 1.8 a resolution
To be Published
|
|
2HAL
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | Hepatitis A Protease 3C, N-ACETYL-LEUCYL-PHENYLALANYL-PHENYLALANYL-GLUTAMATE-FLUOROMETHYLKETONE INHIBITOR, N-[(BENZYLOXY)CARBONYL]-L-ALANINE | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
J.Mol.Biol., 361, 2006
|
|
1Y0H
 
 | | Structure of Rv0793 from Mycobacterium tuberculosis | | Descriptor: | ACETATE ION, hypothetical protein Rv0793 | | Authors: | Lemieux, M.J, Ference, C, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-15 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Rv0793, a hypothetical monooxygenase from M. tuberculosis
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
1PPK
 
 | |
1YUY
 
 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE GENOTYPE 2a | | Descriptor: | RNA-Dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1YVX
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[ISOPROPYL(4-METHYLBENZOYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1YV2
 
 | | Hepatitis C virus NS5B RNA-dependent RNA Polymerase genotype 2a | | Descriptor: | GLYCEROL, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the RNA-dependent RNA Polymerase Genotype 2a of Hepatitis C Virus Reveal Two Conformations and Suggest Mechanisms of Inhibition by Non-nucleoside Inhibitors
J.Biol.Chem., 280, 2005
|
|
1YVZ
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[(2,4-DICHLOROBENZOYL)(ISOPROPYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
2A4O
 
 | | Dual modes of modification of Hepatitis A virus 3C protease by a serine derived beta-lactone: selective crytstallization and high resolution structure of the His102 adduct | | Descriptor: | ACETYL GROUP, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, PHENYLALANINE AMIDE, ... | | Authors: | Yin, J, Bergmann, E.M, Cherney, M.M, Lall, M.S, Jain, R.P, Vederas, J.C, James, M.N.G. | | Deposit date: | 2005-06-29 | | Release date: | 2005-12-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-derived beta-Lactone: Selective Crystallization and Formation of a Functional Catalytic Triad in the Active Site
J.MOL.BIOL., 354, 2005
|
|
3EI9
 
 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with L-Glu: External aldimine form | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-glutamic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EI8
 
 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with LL-DAP: External aldimine form | | Descriptor: | (2S,6S)-2-amino-6-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}heptanedioic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
