3KNQ
 
 | |
8CPQ
 
 | |
5JCQ
 
 | |
5CGB
 
 | | Crystal structure of FimH in complex with heptyl alpha-D-septanoside | | Descriptor: | (6R)-1,6-anhydro-2-O-heptyl-6-(hydroxymethyl)-D-galactitol, Protein FimH, SULFATE ION | | Authors: | Jakob, R.P, Preston, R.P, Zihlmann, P, Fiege, B, Sager, C.P, Vannam, R, Rabbani, S, Zalewski, A, Maier, T, Ernst, B, Peczuh, M. | | Deposit date: | 2015-07-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The price of flexibility - a case study on septanoses as pyranose mimetics.
Chem Sci, 9, 2018
|
|
3DGS
 
 | |
7P1C
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin B | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein assembly factor BamA, TRP-ASN-UX8-THR-LYS-ARG-PHE | | Authors: | Jakob, R.P, Modaresi, S.M, Hiller, S, Maier, T. | | Deposit date: | 2021-07-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutasynthetic Production and Antimicrobial Characterization of Darobactin Analogs.
Microbiol Spectr, 9, 2021
|
|
7AYN
 
 | | Crystal structure of the lectin domain of the FimH variant Arg98Ala, in complex with Methyl 3-chloro-4-D-mannopyranosyloxy-3-biphenylcarboxylate | | Descriptor: | 1,2-ETHANEDIOL, Type 1 fimbrin D-mannose specific adhesin, methyl 3-[3-chloranyl-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-phenyl]benzoate | | Authors: | Jakob, R.P, Tomasic, T, Rabbani, S, Reisner, A, Jakopin, Z, Maier, T, Ernst, B, Anderluh, M. | | Deposit date: | 2020-11-12 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Does targeting Arg98 of FimH lead to high affinity antagonists?
Eur.J.Med.Chem., 211, 2020
|
|
7AQS
 
 | | Crystal structure of E. coli DPS in space group P1 | | Descriptor: | DNA protection during starvation protein, FE (III) ION | | Authors: | Jakob, R.P, Pipercevic, J, Righetto, R, Goldie, K, Stahlberg, H, Maier, T, Hiller, S. | | Deposit date: | 2020-10-22 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a Dps contamination in Mitomycin-C-induced expression of Colicin Ia.
Biochim Biophys Acta Biomembr, 1863, 2021
|
|
7R1V
 
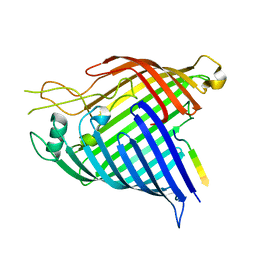 | |
7R1W
 
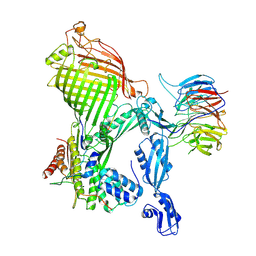 | | E. coli BAM complex (BamABCDE) bound to dynobactin A | | Descriptor: | Dynobactin A, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Hiller, S, Maier, T. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Computational identification of a systemic antibiotic for gram-negative bacteria.
Nat Microbiol, 7, 2022
|
|
5OOW
 
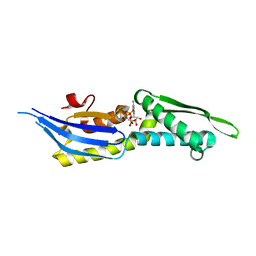 | | Crystal structure of lobe II from the nucleotide binding domain of DnaK in complex with AMPPCP | | Descriptor: | Chaperone protein DnaK, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Jakob, R.P, Bauer, D, Meinhold, S, Stigler, J, Merkel, U, Maier, T, Rief, M, Zoldak, G. | | Deposit date: | 2017-08-09 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A folding nucleus and minimal ATP binding domain of Hsp70 identified by single-molecule force spectroscopy.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MCA
 
 | | Crystal structure of FimH-LD R60P variant in the apo state | | Descriptor: | Protein FimH, SULFATE ION | | Authors: | Jakob, R.P, Rabbani, S, Ernst, B, Maier, T. | | Deposit date: | 2016-11-09 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Conformational switch of the bacterial adhesin FimH in the absence of the regulatory domain: Engineering a minimalistic allosteric system.
J. Biol. Chem., 293, 2018
|
|
5MUC
 
 | | Crystal structure of the FimH lectin domain in complex with 1,5-Anhydromannitol | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, Protein FimH | | Authors: | Jakob, R.P, Rabbani, S, Ernst, B, Maier, T. | | Deposit date: | 2017-01-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | KinITC-One Method Supports both Thermodynamic and Kinetic SARs as Exemplified on FimH Antagonists.
Chemistry, 24, 2018
|
|
8AS4
 
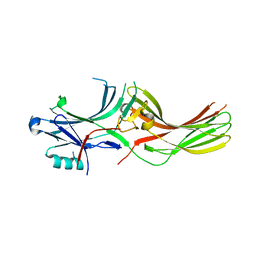 | |
8QEX
 
 | |
8R5M
 
 | | E-selectin complexed with glycomimetic ligand DS0567 | | Descriptor: | (2~{S})-3-cyclohexyl-2-[(2~{R},3~{R},4~{S},5~{S},6~{R})-2-[(1~{R},2~{R},3~{S})-3-ethyl-2-[(2~{S},3~{S},4~{R},5~{S},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-cyclohexyl]oxy-6-(hydroxymethyl)-5-oxidanyl-3-(phenylcarbonyloxy)oxan-4-yl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jakob, R.P, Ernst, B, Maier, T. | | Deposit date: | 2023-11-17 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Analogues of the pan-selectin antagonist rivipansel (GMI-1070).
Eur.J.Med.Chem., 272, 2024
|
|
8R5L
 
 | | E-selectin complexed with glycomimetic ligand BW850 | | Descriptor: | (2~{S})-3-cyclohexyl-2-[(2~{R},3~{R},4~{S},5~{S},6~{R})-2-[(1~{R},2~{R},3~{S},5~{R})-3-ethyl-2-[(2~{S},3~{S},4~{R},5~{S},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-[3-[1-[3-oxidanylidene-3-[(3,6,8-trisulfonaphthalen-1-yl)amino]propyl]-1,2,3-triazol-4-yl]propylcarbamoyl]cyclohexyl]oxy-6-(hydroxymethyl)-5-oxidanyl-3-(phenylcarbonyloxy)oxan-4-yl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jakob, R.P, Ernst, B, Maier, T. | | Deposit date: | 2023-11-17 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Analogues of the pan-selectin antagonist rivipansel (GMI-1070).
Eur.J.Med.Chem., 272, 2024
|
|
7NL7
 
 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | 3-Aminopropyl 2-deoxy-2-(4-phenyl-1,2,3-triazol-1-yl)-alpha-D-mannopyranoside, CALCIUM ION, DC-SIGN, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
7NL6
 
 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | CALCIUM ION, DC-SIGN, CRD domain, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
7NRF
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 2) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRE
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 1) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRI
 
 | | Structure of the darobactin-bound E. coli BAM complex (BamABCDE) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
4EO1
 
 | | crystal structure of the TolA binding domain from the filamentous phage IKe | | Descriptor: | Attachment protein G3P, MAGNESIUM ION | | Authors: | Jakob, R.P, Geitner, A.J, Weininger, U, Balbach, J, Dobbek, H, Schmid, F.X. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and energetic basis of infection by the filamentous bacteriophage IKe.
Mol.Microbiol., 84, 2012
|
|
6HP1
 
 | | Crystal Structure of the O-Methyltransferase from the trans-AT PKS multienzyme C0ZGQ3 of Brevibacillus brevis | | Descriptor: | ETHYL MERCURY ION, Putative polyketide synthase | | Authors: | Jakob, R.P, Felber, P, Demyanenko, Y, Delbart, F, Maier, T. | | Deposit date: | 2018-09-19 | | Release date: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of an O-Methyltransferase from the trans-AT PKS biosynthesis pathway
To Be Published
|
|
4EO0
 
 | | crystal structure of the pilus binding domain of the filamentous phage IKe | | Descriptor: | Attachment protein G3P | | Authors: | Jakob, R.P, Geitner, A.J, Weininger, U, Balbach, J, Dobbek, H, Schmid, F.X. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and energetic basis of infection by the filamentous bacteriophage IKe.
Mol.Microbiol., 84, 2012
|
|
