6QPF
 
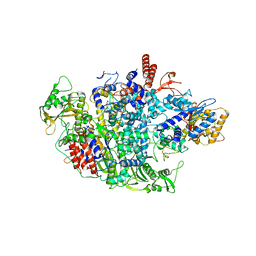 | | Influenza A virus Polymerase Heterotrimer A/duck/Fujian/01/2002(H5N1) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Fan, H.T, Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.634 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QPG
 
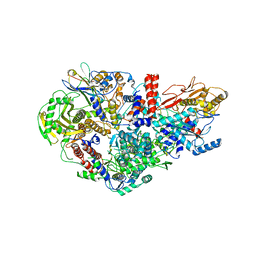 | | Influenza A virus Polymerase Heterotrimer A/nt/60/1968(H3N2) in complex with Nanobody NB8205 | | Descriptor: | Nanobody NB8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Fan, H.T, Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QX8
 
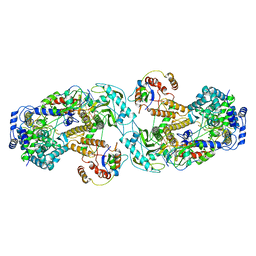 | | Influenza A virus (A/NT/60/1968) polymerase dimer of heterotrimer in complex with 5' cRNA promoter | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(P*AP*GP*CP*AP*AP*AP*AP*GP*CP*AP*GP*A)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QXE
 
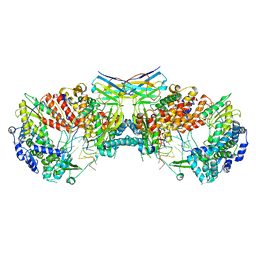 | | Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
4LT2
 
 | | HEWL co-crystallized with Carboplatin in non-NaCl conditions: crystal 2 processed using the EVAL software package | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carboplatin binding to a model protein in non-NaCl conditions to eliminate partial conversion to cisplatin, and the use of different criteria to choose the resolution limit
To be Published
|
|
6QX3
 
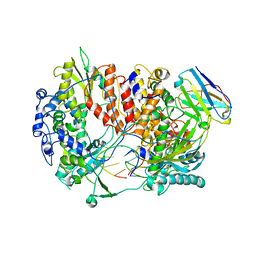 | | Influenza A virus (A/NT/60/1968) polymerase Hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
5MIL
 
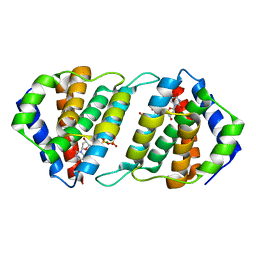 | | Pirating conserved phage mechanisms promotes promiscuous staphylococcal pathogenicity islands transfer. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase family protein, MAGNESIUM ION | | Authors: | Donderis, J, Marina, A, Penades, J.R. | | Deposit date: | 2016-11-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pirating conserved phage mechanisms promotes promiscuous staphylococcal pathogenicity island transfer.
Elife, 6, 2017
|
|
3NGG
 
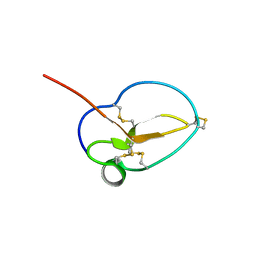 | | X-ray Structure of Omwaprin | | Descriptor: | Omwaprin-a | | Authors: | Banigan, J.R, Mandal, K, Sawaya, M.R, Thammavongsa, V, Hendrickx, A.P.A, Schneewind, O, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2010-06-11 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Determination of the X-ray structure of the snake venom protein omwaprin by total chemical synthesis and racemic protein crystallography.
Protein Sci., 19, 2010
|
|
3NT1
 
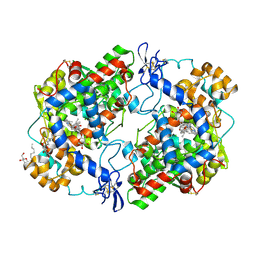 | | High resolution structure of naproxen:COX-2 complex. | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duggan, K.C, Musee, J, Walters, M.J, Harp, J.M, Kiefer, J.R, Oates, J.A, Marnett, L.J. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen.
J.Biol.Chem., 285, 2010
|
|
3NTB
 
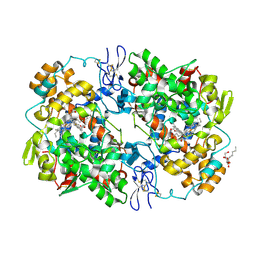 | | Structure of 6-methylthio naproxen analog bound to mCOX-2. | | Descriptor: | (2S)-2-[6-(methylsulfanyl)naphthalen-2-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duggan, K.C, Musee, J, Walters, M.J, Harp, J.M, Kiefer, J.R, Oates, J.A, Marnett, L.J. | | Deposit date: | 2010-07-03 | | Release date: | 2010-09-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen.
J.Biol.Chem., 285, 2010
|
|
1UA0
 
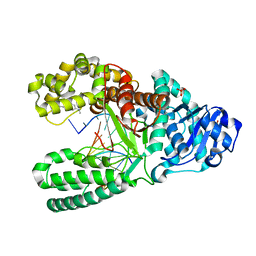 | | Aminofluorene DNA adduct at the pre-insertion site of a DNA polymerase | | Descriptor: | 2-AMINOFLUORENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Kiefer, J.R, Becherel, O.J, Fuchs, R.P.P, Beese, L.S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase
J.Biol.Chem., 279, 2004
|
|
3NPL
 
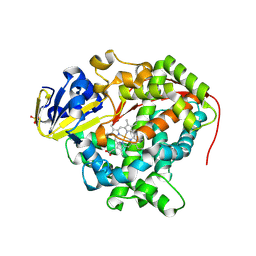 | | Structure of Ru(bpy)2(A-Phen)(K97C) P450 BM3 heme domain, a ruthenium modified P450 BM3 mutant | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Ener, M, Lee, Y.-T, Goodin, D.B, Winkler, J.R, Gray, H.B, Cheruzel, L. | | Deposit date: | 2010-06-28 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Ru(bpy)2(A-Phen)(K97C) P450 BM3 heme domain, a ruthenium modified P450 BM3 mutant
To be Published
|
|
1UA1
 
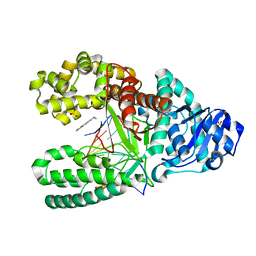 | | Structure of aminofluorene adduct paired opposite cytosine at the polymerase active site. | | Descriptor: | 2-AMINOFLUORENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Kiefer, J.R, Becherel, O.J, Fuchs, R.P.P, Beese, L.S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase
J.Biol.Chem., 279, 2004
|
|
3O8D
 
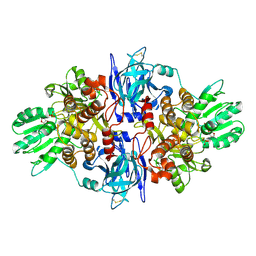 | |
3O2X
 
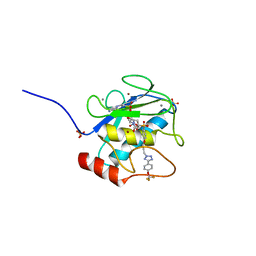 | | MMP-13 in complex with selective tetrazole core inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Collagenase 3, ... | | Authors: | Kiefer, J.R, Barta, T.E, Becker, D.P, Mathis, K.J, Williams, J. | | Deposit date: | 2010-07-22 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MMP-13 in complex with selective tetrazole core inhibitor
To be Published
|
|
3O8C
 
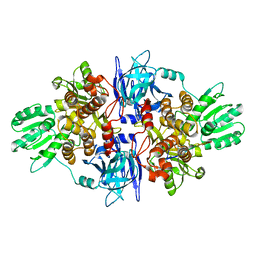 | |
6T64
 
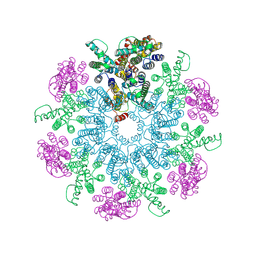 | | A model of the EIAV CA-SP hexamer (C6) from Gag-deltaMA spheres assembled at pH6 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
6T61
 
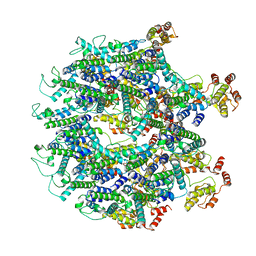 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH8 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
6T63
 
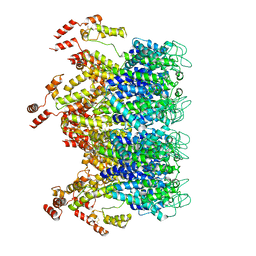 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH6 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
5JN2
 
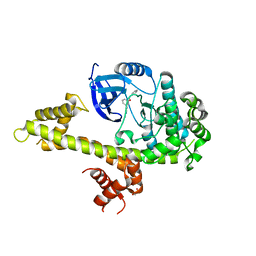 | | Crystal structure of TgCDPK1 bound to NVPACU106 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calmodulin-domain protein kinase 1, ... | | Authors: | El Bakkouri, M, Walker, J.R, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Lovato, D.V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-04-29 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TgCDPK1 bound to NVPACU106
To Be Published
|
|
5JJW
 
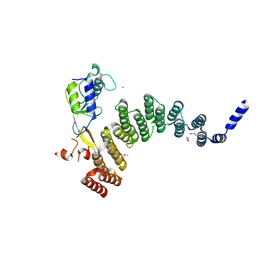 | | Crystal structure of the HAT domain of sart3 in complex with USP15 DUSP-UBL domain | | Descriptor: | 1,2-ETHANEDIOL, Squamous cell carcinoma antigen recognized by T-cells 3, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Zhang, Q, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure of the HAT domain of sart3 in complex with USP15 DUSP-UBL domain
to be published
|
|
5J3U
 
 | | Co-crystal structure of the regulatory domain of Toxoplasma gondii PKA with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, Protein Kinase A | | Authors: | El Bakkouri, M, Walker, J.R, Tempel, W, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Lin, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-31 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Co-crystal structure of the regulatory domain of Toxoplasma gondii PKA with cAMP
To Be Published
|
|
5J7D
 
 | | Computationally Designed Thioredoxin dF106 | | Descriptor: | COPPER (II) ION, Designed Thioredoxin dF106 | | Authors: | Horowitz, S, Johansen, N, Olsen, J.G, Winther, J.R. | | Deposit date: | 2016-04-06 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational Redesign of Thioredoxin Is Hypersensitive toward Minor Conformational Changes in the Backbone Template.
J.Mol.Biol., 428, 2016
|
|
5IUT
 
 | | STRUCTURE OF P450 2B4 F202W MUTANT | | Descriptor: | 3,6,9,12,15,18-hexaoxahexacosan-1-ol, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jang, H.-H, Halpert, J.R, Shah, M.B. | | Deposit date: | 2016-03-18 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Effect of detergent binding on cytochrome P450 2B4 structure as analyzed by X-ray crystallography and deuterium-exchange mass spectrometry.
Biophys.Chem., 216, 2016
|
|
5J77
 
 | |
