3UDG
 
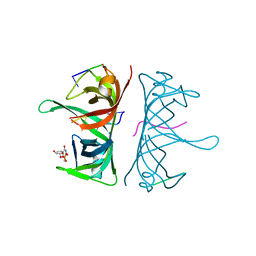 | | Structure of Deinococcus radiodurans SSB bound to ssDNA | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3', Single-stranded DNA-binding protein, THYMIDINE-5'-PHOSPHATE | | Authors: | George, N.P, Ngo, K.V, Chitteni-Patu, S, Norais, C.A, Battista, J.R, Cox, M.M, Keck, J.L. | | Deposit date: | 2011-10-28 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Cellular Dynamics of Deinococcus radiodurans Single-stranded DNA (ssDNA)-binding Protein (SSB)-DNA Complexes.
J.Biol.Chem., 287, 2012
|
|
3UDL
 
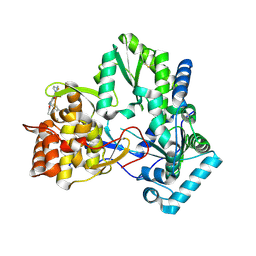 | | 3-heterocyclyl quinolone bound to HCV NS5B | | Descriptor: | 3-(5-benzyl-1,2,4-oxadiazol-3-yl)-6-fluoro-1-[2-fluoro-4-(trifluoromethyl)benzyl]-7-(4-methylpiperazin-1-yl)quinolin-4(1H)-one, HCV NS5B polymerase | | Authors: | Somoza, J.R. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Quinolones as HCV NS5B polymerase inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3UEK
 
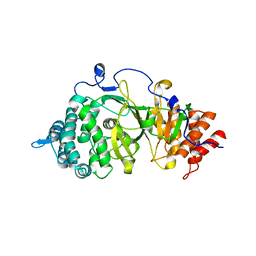 | | Crystal structure of the catalytic domain of rat poly (ADP-ribose) glycohydrolase | | Descriptor: | Poly(ADP-ribose) glycohydrolase | | Authors: | Kim, I.K, Kiefer, J.R, Stegemann, R.A, Classen, S, Tainer, J.A, Ellenberger, T. | | Deposit date: | 2011-10-30 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UEL
 
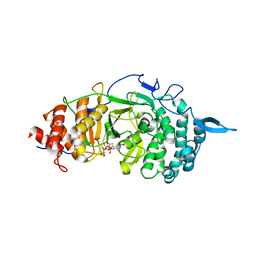 | | Crystal structure of the catalytic domain of rat poly (ADP-ribose) glycohydrolase bound to ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Poly(ADP-ribose) glycohydrolase | | Authors: | Kim, I.K, Kiefer, J.R, Stegemann, R.A, Classen, S, Tainer, J.A, Ellenberger, T. | | Deposit date: | 2011-10-30 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UIN
 
 | | Complex between human RanGAP1-SUMO2, UBC9 and the IR1 domain from RanBP2 | | Descriptor: | E3 SUMO-protein ligase RanBP2, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Gareau, J.R, Reverter, D, Lima, C.D. | | Deposit date: | 2011-11-05 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
J.Biol.Chem., 287, 2012
|
|
3UWK
 
 | | Structure Guided Development of Novel Thymidine Mimetics targeting Pseudomonas aeruginosa Thymidylate Kinase: from Hit to Lead Generation | | Descriptor: | 1-methyl-6-phenyl-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, MAGNESIUM ION, Thymidylate kinase | | Authors: | Choi, J.Y, Plummer, M.S, Starr, J, Desbonnet, C.R, Soutter, H.H, Chang, J, Miller, J.R, Dillman, K, Miller, A.A, Roush, W.R. | | Deposit date: | 2011-12-02 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure guided development of novel thymidine mimetics targeting Pseudomonas aeruginosa thymidylate kinase: from hit to lead generation.
J.Med.Chem., 55, 2012
|
|
3UIP
 
 | | Complex between human RanGAP1-SUMO1, UBC9 and the IR1 domain from RanBP2 containing IR2 Motif II | | Descriptor: | E3 SUMO-protein ligase RanBP2, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Gareau, J.R, Reverter, D, Lima, C.D. | | Deposit date: | 2011-11-05 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
J.Biol.Chem., 287, 2012
|
|
3UWO
 
 | | Structure Guided Development of Novel Thymidine Mimetics targeting Pseudomonas aeruginosa Thymidylate Kinase: from Hit to Lead Generation | | Descriptor: | 3-(1-methyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-6-yl)benzamide, Thymidylate kinase | | Authors: | Choi, J.Y, Plummer, M.S, Starr, J, Desbonnet, C.R, Soutter, H.H, Chang, J, Miller, J.R, Dillman, K, Miller, A.A, Roush, W.R. | | Deposit date: | 2011-12-02 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure guided development of novel thymidine mimetics targeting Pseudomonas aeruginosa thymidylate kinase: from hit to lead generation.
J.Med.Chem., 55, 2012
|
|
1MOL
 
 | |
4W8R
 
 | |
2FN2
 
 | | SOLUTION NMR STRUCTURE OF THE GLYCOSYLATED SECOND TYPE TWO MODULE OF FIBRONECTIN, 20 STRUCTURES | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FIBRONECTIN | | Authors: | Sticht, H, Pickford, A.R, Potts, J.R, Campbell, I.D. | | Deposit date: | 1997-08-06 | | Release date: | 1998-09-16 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the glycosylated second type 2 module of fibronectin.
J.Mol.Biol., 276, 1998
|
|
4OCS
 
 | | Crystal structure of human Fab CAP256-VRC26.10, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.10 heavy chain, CAP256-VRC26.10 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4OPF
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS8 | | Descriptor: | NRPS/PKS | | Authors: | Osipiuk, J, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4OO2
 
 | | Streptomyces globisporus C-1027 FAD dependent (S)-3-chloro-β-tyrosine-S-SgcC2 C-5 hydroxylase SgcC apo form | | Descriptor: | CALCIUM ION, Chlorophenol-4-monooxygenase, GLYCEROL | | Authors: | Cao, H, Xu, W, Bingman, C.A, Lohman, J.R, Yennamalli, R, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-01-29 | | Release date: | 2014-02-12 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
4OWC
 
 | | PtI6 binding to HEWL | | Descriptor: | CHLORIDE ION, IODIDE ION, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Starkey, V.L, Lamplough, L, Kaenket, S, Helliwell, J.R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The binding of platinum hexahalides (Cl, Br and I) to hen egg-white lysozyme and the chemical transformation of the PtI6 octahedral complex to a PtI3 moiety bound to His15.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4OCW
 
 | | Crystal structure of human Fab CAP256-VRC26.06, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.06 heavy chain, CAP256-VRC26.06 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
5K9K
 
 | | Crystal structure of multidonor HV6-1-class broadly neutralizing Influenza A antibody 56.a.09 in complex with Hemagglutinin Hong Kong 1968. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 56.a.09 Heavy chain, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-31 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
5KAQ
 
 | | Crystal structure of broadly neutralizing Influenza A antibody 31.a.83 in complex with Hemagglutinin Hong Kong 1968. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 31.A.83 FAB HEAVY CHAIN, ANTIBODY 31.A.83 FAB LIGHT CHAIN, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
5K9O
 
 | | Crystal structure of multidonor HV1-18+HD3-9 class broadly neutralizing Influenza A antibody 31.b.09 in complex with Hemagglutinin H1 A/California/04/2009 | | Descriptor: | 31.b.09 Heavy Fv, 31.b.09 Light Fv, Hemagglutinin, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-06-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.387 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
5K9Q
 
 | | Crystal structure of multidonor HV1-18-class broadly neutralizing Influenza A antibody 16.a.26 in complex with A/Hong Kong/1-4-MA21-1/1968 (H3N2) Hemagglutinin | | Descriptor: | 16.a.26 Heavy chain, 16.a.26 Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell(Cambridge,Mass.), 166, 2016
|
|
1M6D
 
 | | Crystal structure of human cathepsin F | | Descriptor: | 4-MORPHOLIN-4-YL-PIPERIDINE-1-CARBOXYLIC ACID [1-(3-BENZENESULFONYL-1-PROPYL-ALLYLCARBAMOYL)-2-PHENYLETHYL]-AMIDE, Cathepsin F | | Authors: | Somoza, J.R, Palmer, J.T, Ho, J.D. | | Deposit date: | 2002-07-15 | | Release date: | 2003-07-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of human cathepsin F and its implications for the development of novel immunomodulators
J.Mol.Biol., 322, 2002
|
|
1M4X
 
 | | PBCV-1 virus capsid, quasi-atomic model | | Descriptor: | PBCV-1 virus capsid | | Authors: | Nandhagopal, N, Simpson, A.A, Gurnon, J.R, Yan, X, Baker, T.S, Graves, M.V, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2002-07-05 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | The Structure and Evolution of the Major Capsid Protein of a Large,
Lipid containing, DNA virus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1M6E
 
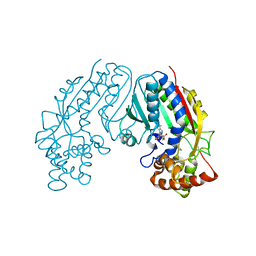 | | CRYSTAL STRUCTURE OF SALICYLIC ACID CARBOXYL METHYLTRANSFERASE (SAMT) | | Descriptor: | 2-HYDROXYBENZOIC ACID, LUTETIUM (III) ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zubieta, C, Ross, J.R, Koscheski, P, Yang, Y, Pichersky, E, Noel, J.P. | | Deposit date: | 2002-07-16 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Substrate Recognition in The Salicylic Acid Carboxyl Methyltransferase Family
Plant Cell, 15, 2003
|
|
1MOZ
 
 | | ADP-ribosylation factor-like 1 (ARL1) from Saccharomyces cerevisiae | | Descriptor: | ADP-ribosylation factor-like protein 1, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Amor, J.C, Horton, J.R, Zhu, X, Wang, Y, Sullards, C, Ringe, D, Cheng, X, Kahn, R.A. | | Deposit date: | 2002-09-10 | | Release date: | 2002-10-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structures of Yeast ARF2 and ARL1:
DISTINCT ROLES FOR THE N TERMINUS IN THE STRUCTURE
AND FUNCTION OF ARF FAMILY GTPases
J.Biol.Chem., 276, 2001
|
|
1LV3
 
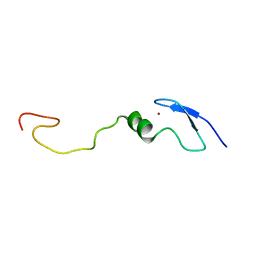 | | Solution NMR Structure of Zinc Finger Protein yacG from Escherichia coli. Northeast Structural Genomics Consortium Target ET92. | | Descriptor: | HYPOTHETICAL PROTEIN YacG, ZINC ION | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Semesi, A, Edwards, A.M, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-05-24 | | Release date: | 2002-09-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Escherichia coli protein YacG: a novel sequence motif in the zinc-finger family of proteins.
Proteins, 49, 2002
|
|
