5EJD
 
 | | The crystal structure of holo T3CT | | Descriptor: | 4'-PHOSPHOPANTETHEINE, GLYCEROL, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
5EGF
 
 | | The crystal structure of SeMet-CT | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-10-27 | | Release date: | 2016-10-19 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
5EPK
 
 | | Crystal Structure of chromodomain of CBX2 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 2, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
7O05
 
 | |
7O36
 
 | |
7O35
 
 | |
5W9Q
 
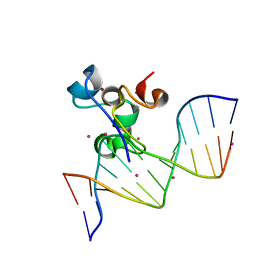 | | Zinc finger region of MBD1 in complex with CpG DNA | | Descriptor: | Methyl-CpG-binding domain protein 1, UNKNOWN ATOM OR ION, ZINC ION, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-23 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
5EPL
 
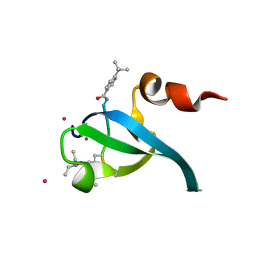 | | Crystal Structure of chromodomain of CBX4 in complex with inhibitor UNC3866 | | Descriptor: | E3 SUMO-protein ligase CBX4, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
9GG1
 
 | | P301T type I tau filaments from human brain | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
9GG6
 
 | | P301T type II tau filaments from human brain | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-13 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
9GG0
 
 | | P301L tau filaments from human brain | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
10MH
 
 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH ADOHCY AND HEMIMETHYLATED DNA CONTAINING 5,6-DIHYDRO-5-AZACYTOSINE AT THE TARGET | | Descriptor: | DNA (5'-D(P*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*AP*GP*5NCP*GP*CP*AP*TP*GP*G)-3'), PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), ... | | Authors: | Sheikhnejad, G, Brank, A, Christman, J.K, Goddard, A, Alvarez, E, Ford Junior, H, Marquez, V.E, Marasco, C.J, Sufrin, J.R, O'Gara, M, Cheng, X. | | Deposit date: | 1998-08-10 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of inhibition of DNA (cytosine C5)-methyltransferases by oligodeoxyribonucleotides containing 5,6-dihydro-5-azacytosine.
J.Mol.Biol., 285, 1999
|
|
1AH5
 
 | | REDUCED FORM SELENOMETHIONINE-LABELLED HYDROXYMETHYLBILANE SYNTHASE DETERMINED BY MAD | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Harrop, S.J, Cassetta, A. | | Deposit date: | 1997-04-13 | | Release date: | 1997-10-15 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of the structure of seleno-methionine-labelled hydroxymethylbilane synthase in its active form by multi-wavelength anomalous dispersion.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
184D
 
 | | SELF-ASSOCIATION OF A DNA LOOP CREATES A QUADRUPLEX: CRYSTAL STRUCTURE OF D(GCATGCT) AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, Zhang, S, Peterson, M.R, Harrop, S.J, Helliwell, J.R, Cruse, W.B.T, Langlois D'Estaintot, B, Kennard, O, Brown, T, Hunter, W.N. | | Deposit date: | 1994-08-10 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Self-association of a DNA loop creates a quadruplex: crystal structure of d(GCATGCT) at 1.8 A resolution.
Structure, 3, 1995
|
|
1AIU
 
 | | HUMAN THIOREDOXIN (D60N MUTANT, REDUCED FORM) | | Descriptor: | THIOREDOXIN | | Authors: | Andersen, J.F, Gasdaska, J.R, Sanders, D.A.R, Weichsel, A, Powis, G, Montfort, W.R. | | Deposit date: | 1997-04-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human thioredoxin homodimers: regulation by pH, role of aspartate 60, and crystal structure of the aspartate 60 --> asparagine mutant.
Biochemistry, 36, 1997
|
|
1AH2
 
 | | SERINE PROTEASE PB92 FROM BACILLUS ALCALOPHILUS, NMR, 18 STRUCTURES | | Descriptor: | SERINE PROTEASE PB92 | | Authors: | Boelens, R, Schipper, D, Martin, J.R, Karimi-Nejad, Y, Mulder, F, Zwan, J.V.D, Mariani, M. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of serine protease PB92 from Bacillus alcalophilus presents a rigid fold with a flexible substrate-binding site.
Structure, 5, 1997
|
|
1A8A
 
 | | RAT ANNEXIN V COMPLEXED WITH GLYCEROPHOSPHOSERINE | | Descriptor: | ANNEXIN V, CALCIUM ION, L-ALPHA-GLYCEROPHOSPHORYLSERINE | | Authors: | Swairjo, M.A, Concha, N.O, Kaetzel, M.A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 1998-03-23 | | Release date: | 1998-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ca(2+)-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V.
Nat.Struct.Biol., 2, 1995
|
|
1A8B
 
 | | RAT ANNEXIN V COMPLEXED WITH GLYCEROPHOSPHOETHANOLAMINE | | Descriptor: | ANNEXIN V, CALCIUM ION, L-ALPHA-GLYCEROPHOSPHORYLETHANOLAMINE | | Authors: | Swairjo, M.A, Concha, N.O, Kaetzel, M.A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 1998-03-23 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ca(2+)-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V.
Nat.Struct.Biol., 2, 1995
|
|
1AVO
 
 | | PROTEASOME ACTIVATOR REG(ALPHA) | | Descriptor: | 11S REGULATOR | | Authors: | Hill, C.P, Knowlton, J.R. | | Deposit date: | 1997-09-18 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the proteasome activator REGalpha (PA28alpha).
Nature, 390, 1997
|
|
7P7W
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P7I
 
 | | Native structure of N-acetylglucosamine kinase from Plesiomonas shigelloides | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9L
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9P
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine and AMP-PNP inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7PA1
 
 | | Structure of N-acetylglucosamine kinase from Plesiomonas shigelloides in complex with AMP-PNP in the absence of N-acetylglucoseamine substrate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9Y
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
