8SA6
 
 | | apo form of adenosylcobalamin riboswitch dimer | | Descriptor: | apo form of adenosylcobalamin riboswitch dimer | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA5
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 4 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 4 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SSS
 
 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) Complexed with 23mer | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8STI
 
 | | human STING with agonist XMT-1616 | | Descriptor: | 3-[(2E)-4-{5-carbamoyl-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-7-(3-hydroxypropoxy)-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-3H-imidazo[4,5-b]pyridine-6-carboxamide, Stimulator of interferon genes protein | | Authors: | Duvall, J.R, Bukhalid, R.A. | | Deposit date: | 2023-05-10 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and Optimization of a STING Agonist Platform for Application in Antibody Drug Conjugates.
J.Med.Chem., 66, 2023
|
|
8SSQ
 
 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-4 | | Descriptor: | DNA (35-MER) Strand 2, DNA (35-MER) Strand I, SODIUM ION, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSR
 
 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-20 | | Descriptor: | DNA (35-MER) Strand I, DNA (35-MER) Strand II, SODIUM ION, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SA4
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 3 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 3 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8S9O
 
 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - P1 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9N
 
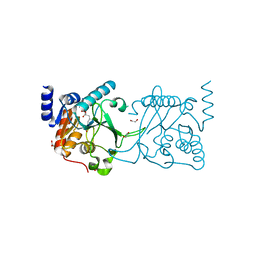 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - C2 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8SO0
 
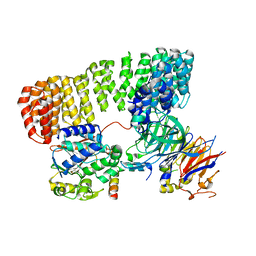 | | Cryo-EM structure of the PP2A:B55-FAM122A complex | | Descriptor: | FE (III) ION, PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-04-28 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.79961 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
8BCQ
 
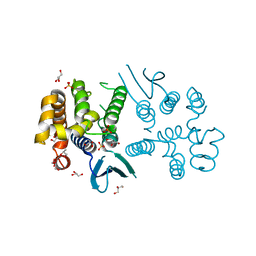 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (native crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION | | Authors: | Benas, P, Jaramillo Ponce, J.R, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
7AYY
 
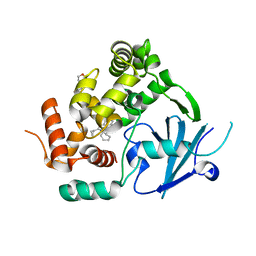 | | Structure of the human 8-oxoguanine DNA Glycosylase hOGG1 in complex with activator TH10785 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Davies, J.R, Stenmark, P. | | Deposit date: | 2020-11-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function.
Science, 376, 2022
|
|
7AYZ
 
 | |
7AZ0
 
 | |
5NZ2
 
 | | Twist and induce: Dissecting the link between the enzymatic activity and the SaPI inducing capacity of the phage 80 dUTPase. D95E mutant from dUTPase 80alpha phage. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase, MAGNESIUM ION | | Authors: | Alite, C, Humphrey, S, Donderis, J, Maiques, E, Ciges-Tomas, J.R, Penades, J.R, Marina, A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dissecting the link between the enzymatic activity and the SaPI inducing capacity of the phage 80 alpha dUTPase.
Sci Rep, 7, 2017
|
|
5NYZ
 
 | | Twist and induce: Dissecting the link between the enzymatic activity and the SaPI inducing capacity of the phage 80 dUTPase. D95E mutant from dUTPase 80alpha phage. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase, MAGNESIUM ION | | Authors: | Alite, C, Humphrey, S, Donderis, J, Maiques, E, Ciges-Tomas, J.R, Penades, J.R, Marina, A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dissecting the link between the enzymatic activity and the SaPI inducing capacity of the phage 80 alpha dUTPase.
Sci Rep, 7, 2017
|
|
8VDI
 
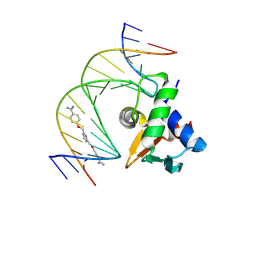 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGAGGAAGTGGG) in Ternary Complex with DB2447 | | Descriptor: | 4,4'-[pyridine-2,6-diylbis(methyleneoxy)]di(benzene-1-carboximidamide), DNA (5'-D(*AP*AP*TP*AP*AP*GP*AP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*TP*CP*TP*TP*AP*T)-3'), ... | | Authors: | Terrell, J.R, Ogbonna, E.N, Poon, G.M.K, Wilson, W.D. | | Deposit date: | 2023-12-15 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Single GC base pair recognition by a heterocyclic diamidine: structures, affinities, and dynamics.
Rsc Adv, 14, 2024
|
|
8VPG
 
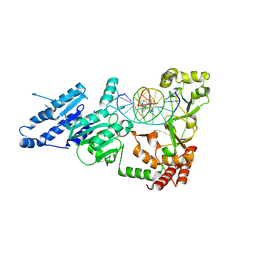 | |
8VPI
 
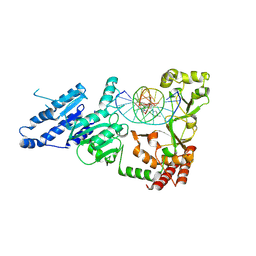 | |
8VPH
 
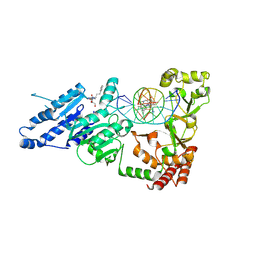 | |
8VDH
 
 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAGAAGGAAGTGGG) in Ternary Complex with DB2447 | | Descriptor: | 4,4'-[pyridine-2,6-diylbis(methyleneoxy)]di(benzene-1-carboximidamide), DNA (5'-D(*AP*AP*TP*AP*GP*AP*AP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*TP*TP*CP*TP*AP*T)-3'), ... | | Authors: | Terrell, J.R, Ogbonna, E.N, Poon, G.M.K, Wilson, W.D. | | Deposit date: | 2023-12-15 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Single GC base pair recognition by a heterocyclic diamidine: structures, affinities, and dynamics.
Rsc Adv, 14, 2024
|
|
8V9N
 
 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAGAAGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*GP*AP*AP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*TP*TP*CP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Ogbonna, E.N, Poon, G.M.K, Wilson, W.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Single GC base pair recognition by a heterocyclic diamidine: structures, affinities, and dynamics.
Rsc Adv, 14, 2024
|
|
8V4T
 
 | |
8VIU
 
 | |
