3H9R
 
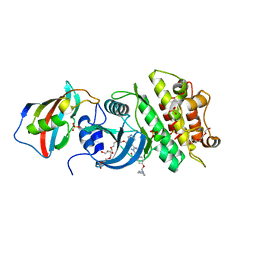 | | Crystal structure of the kinase domain of type I activin receptor (ACVR1) in complex with FKBP12 and dorsomorphin | | Descriptor: | 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine, Activin receptor type-1, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Chaikuad, A, Alfano, I, Shrestha, B, Muniz, J.R.C, Petrie, K, Fedorov, O, Phillips, C, Bishop, S, Mahajan, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Bone Morphogenetic Protein Receptor ALK2 and Implications for Fibrodysplasia Ossificans Progressiva.
J.Biol.Chem., 287, 2012
|
|
1J40
 
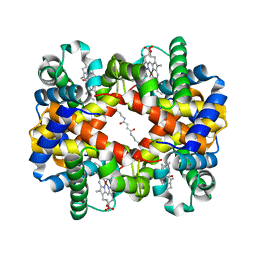 | | Direct observation of photolysis-induced tertiary structural changes in human haemoglobin; Crystal structure of alpha(Ni)-beta(Fe-CO) hemoglobin (laser unphotolysed) | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin alpha Chain, ... | | Authors: | Adachi, S, Park, S.-Y, Tame, J.R.H, Shiro, Y, Shibayama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-21 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Direct observation of photolysis-induced tertiary structural changes in hemoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3P1M
 
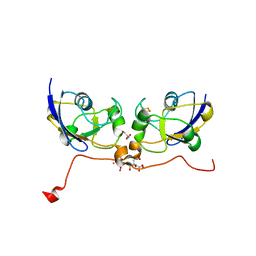 | | Crystal structure of human ferredoxin-1 (FDX1) in complex with iron-sulfur cluster | | Descriptor: | Adrenodoxin, mitochondrial, CITRATE ANION, ... | | Authors: | Chaikuad, A, Johansson, C, Krojer, T, Yue, W.W, Phillips, C, Bray, J.E, Pike, A.C.W, Muniz, J.R.C, Vollmar, M, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of human ferredoxin-1 (FDX1) in complex with iron-sulfur cluster
To be Published
|
|
1MVH
 
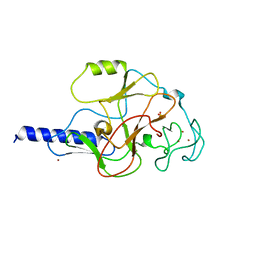 | | structure of the SET domain histone lysine methyltransferase Clr4 | | Descriptor: | Cryptic loci regulator 4, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Min, J.R, Zhang, X, Cheng, X.D, Grewal, S.I.S, Xu, R.-M. | | Deposit date: | 2002-09-25 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the SET domain histone lysine methyltransferase Clr4.
Nat.Struct.Biol., 9, 2002
|
|
1J3Z
 
 | | Direct observation of photolysis-induced tertiary structural changes in human haemoglobin; Crystal structure of alpha(Fe-CO)-beta(Ni) hemoglobin (laser unphotolysed) | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin alpha Chain, ... | | Authors: | Adachi, S, Park, S.-Y, Tame, J.R.H, Shiro, Y, Shibayama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-21 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct observation of photolysis-induced tertiary structural changes in hemoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1IV3
 
 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase (bound form MG atoms) | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, MAGNESIUM ION | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5TEE
 
 | | Crystal structure of Gemin5 WD40 repeats in apo form | | Descriptor: | GLYCEROL, Gem-associated protein 5, SODIUM ION, ... | | Authors: | Chao, X, Tempel, W, Bian, C, Cerovina, T, He, H, Walker, J.R, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-21 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly.
Genes Dev., 30, 2016
|
|
3PRY
 
 | | Crystal structure of the middle domain of human HSP90-beta refined at 2.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Heat shock protein HSP 90-beta, ... | | Authors: | Chaikuad, A, Pilka, E, Sharpe, T.D, Cooper, C.D.O, Phillips, C, Berridge, G, Ayinampudi, V, Fedorov, O, Keates, T, Thangaratnarajah, C, Zimmermann, T, Vollmar, M, Yue, W.W, Che, K.H, Krojer, T, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-30 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the middle domain of human HSP90-beta refined at 2.3 A resolution
To be Published
|
|
3GNO
 
 | | Crystal Structure of a Rice Os3BGlu6 Beta-Glucosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Os03g0212800 protein | | Authors: | Seshadri, S, Akiyama, T, Opassiri, R, Kuaprasert, B, Cairns, J.R.K. | | Deposit date: | 2009-03-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural and enzymatic characterization of Os3BGlu6, a rice {beta}-glucosidase hydrolyzing hydrophobic glycosides and (1->3)- and (1->2)-linked disaccharides.
Plant Physiol., 2009
|
|
3GNP
 
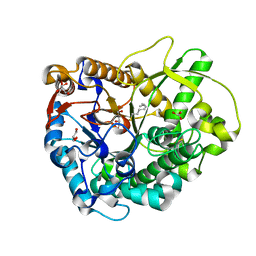 | | Crystal Structure of a Rice Os3BGlu6 Beta-Glucosidase with Octyl-Beta-D-Thio-Glucoside | | Descriptor: | GLYCEROL, Os03g0212800 protein, octyl 1-thio-beta-D-glucopyranoside | | Authors: | Seshadri, S, Akiyama, T, Opassiri, R, Kuaprasert, B, Cairns, J.R.K. | | Deposit date: | 2009-03-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and enzymatic characterization of Os3BGlu6, a rice {beta}-glucosidase hydrolyzing hydrophobic glycosides and (1->3)- and (1->2)-linked disaccharides.
Plant Physiol., 2009
|
|
3PFV
 
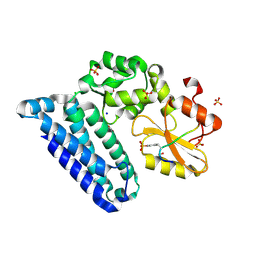 | | Crystal structure of Cbl-b TKB domain in complex with EGFR pY1069 peptide | | Descriptor: | 1,2-ETHANEDIOL, 11-meric peptide from Epidermal growth factor receptor, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Guo, K, Cooper, C.D.O, Ayinampudi, V, Krojer, T, Muniz, J.R.C, Vollmar, M, Canning, P, Gileadi, O, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of Cbl-b TKB domain in complex with EGFR pY1069 peptide
To be Published
|
|
3PS4
 
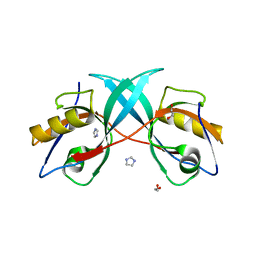 | | PDZ domain from Human microtubule-associated serine/threonine-protein kinase 1 | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Microtubule-associated serine/threonine-protein kinase 1 | | Authors: | Ugochukwu, E, Wang, J, Krojer, T, Muniz, J.R.C, Sethi, R, Pike, A.C.W, Roos, A, Salah, E, Cocking, R, Savitsky, P, Doyle, D.A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-30 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PDZ domain from Human microtubule-associated serine/threonine-protein kinase 1
TO BE PUBLISHED
|
|
1WUB
 
 | | Crystal structure of the polyisoprenoid-binding protein, TT1927b, from Thermus thermophilus HB8 | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, conserved hypothetical protein TT1927b | | Authors: | Handa, N, Idaka, M, Terada, T, Hamana, H, Ishizuka, Y, Park, S.-Y, Tame, J.R.H, Doi-Katayama, Y, Hirota, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-12-03 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a novel polyisoprenoid-binding protein from Thermus thermophilus HB8
Protein Sci., 14, 2005
|
|
3I5P
 
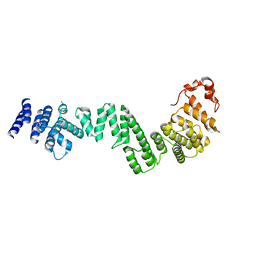 | | Nup170(aa979-1502), S.cerevisiae | | Descriptor: | Nucleoporin NUP170 | | Authors: | Whittle, J.R.R, Schwartz, T.U. | | Deposit date: | 2009-07-06 | | Release date: | 2009-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element.
J.Biol.Chem., 284, 2009
|
|
3IXR
 
 | | Crystal Structure of Xylella fastidiosa PrxQ C47S Mutant | | Descriptor: | Bacterioferritin comigratory protein | | Authors: | Horta, B.B, Oliveira, M.A, Discola, K.F, Cussiol, J.R.R, Netto, L.E.S. | | Deposit date: | 2009-09-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Analysis Indicates that PrxQ, a Cys Based Peroxidase from Xylella fastidiosa, possesses high reactivity towards hydroperoxides
To be Published
|
|
2RKM
 
 | | STRUCTURE OF OPPA COMPLEXED WITH LYS-LYS | | Descriptor: | LYSINE, OLIGO-PEPTIDE BINDING PROTEIN, URANYL (VI) ION | | Authors: | Sleigh, S.H, Tame, J.R.H, Wilkinson, A.J. | | Deposit date: | 1997-03-25 | | Release date: | 1997-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine.
Biochemistry, 36, 1997
|
|
5U4X
 
 | | Coactivator-associated arginine methyltransferase 1 with TP-064 | | Descriptor: | Histone-arginine methyltransferase CARM1, N-methyl-N-[(2-{1-[2-(methylamino)ethyl]piperidin-4-yl}pyridin-4-yl)methyl]-3-phenoxybenzamide, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | DONG, A, ZENG, H, Saikatendu, K.S, STONE, H, WALKER, J.R, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-06 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma.
Oncotarget, 9, 2018
|
|
2C7H
 
 | | Solution NMR structure of the DWNN domain from human RBBP6 | | Descriptor: | RETINOBLASTOMA-BINDING PROTEIN 6, ISOFORM 3 | | Authors: | Pugh, D.J.R, Ab, E, Faro, A, Lutya, P.T, Hoffmann, E, Rees, D.J.G. | | Deposit date: | 2005-11-24 | | Release date: | 2006-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dwnn, a Novel Ubiquitin-Like Domain, Implicates Rbbp6 in Mrna Processing and Ubiquitin-Like Pathways
Bmc Struct.Biol., 6, 2006
|
|
1GTT
 
 | | CRYSTAL STRUCTURE OF HPCE | | Descriptor: | 4-HYDROXYPHENYLACETATE DEGRADATION BIFUNCTIONAL ISOMERASE/DECARBOXYLASE, CALCIUM ION | | Authors: | Tame, J.R.H, Namba, K, Dodson, E.J, Roper, D.I. | | Deposit date: | 2002-01-18 | | Release date: | 2002-03-08 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Hpce, a Bifunctional Decarboxylase/Isomerase with a Multifunctional Fold.
Biochemistry, 41, 2002
|
|
1QIC
 
 | | CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN | | Descriptor: | CALCIUM ION, PROTEIN (STROMELYSIN-1), ZINC ION | | Authors: | Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A.G, Rubin, J.R, Skeean, R.W, White, A.D, Blundell, T.L, Humblet, C, Hupe, D.J, Dhanaraj, V. | | Deposit date: | 1999-06-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity
Protein Sci., 8, 1999
|
|
3Q2F
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with a triazolo ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[3-(1H-imidazol-1-yl)propyl]-5-methyl-4H-1,2,4-triazol-3-yl}-1-methyl-1H-pyrazol-5-amine, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Muniz, J.R.C, Filippakopoulos, P, Picaud, S, Felletar, I, Keates, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-20 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the bromodomain of human BAZ2B in complex with a triazolo ligand
TO BE PUBLISHED
|
|
3P1A
 
 | | Structure of human Membrane-associated Tyrosine- and Threonine-specific cdc2-inhibitory kinase MYT1 (PKMYT1) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Chaikuad, A, Eswaran, J, Fedorov, O, Cooper, C.D.O, Kroeler, T, Vollmar, M, Krojer, T, Berridge, G, Muniz, J.R.C, Pike, A.C.W, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human Membrane-associated Tyrosine- and Threonine-specific cdc2-inhibitory kinase MYT1 (PKMYT1)
To be Published
|
|
6D2L
 
 | | Crystal structure of human CARM1 with (S)-SKI-72 | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-hydroxyphenyl)ethyl]hexanamide, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Hutchinson, A, Seitova, A, LUO, M, CAI, X.C, KE, W, WANG, J, SHI, C, ZHENG, W, LEE, J.P, IBANEZ, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
3II7
 
 | | Crystal structure of the kelch domain of human KLHL7 | | Descriptor: | 1,2-ETHANEDIOL, Kelch-like protein 7 | | Authors: | Chaikuad, A, Thangaratnarajah, C, Cooper, C.D.O, Ugochukwu, E, Muniz, J.R.C, Krojer, T, Sethi, R, Pike, A.C.W, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases.
J.Biol.Chem., 288, 2013
|
|
1SKJ
 
 | | COCRYSTAL STRUCTURE OF UREA-SUBSTITUTED PHOSPHOPEPTIDE COMPLEX | | Descriptor: | 4-[3-CARBOXYMETHYL-3-(4-PHOSPHONOOXY-BENZYL)-UREIDO]-4-[(3-CYCLOHEXYL-PROPYL)-METHYL-CARBAMOYL]BUTYRIC ACID, PP60 V-SRC TYROSINE KINASE TRANSFORMING PROTEIN | | Authors: | Holland, D.R, Rubin, J.R. | | Deposit date: | 1997-09-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and cocrystal structure of a nonpeptide Src SH2 domain ligand.
J.Med.Chem., 40, 1997
|
|
