4IUW
 
 | | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20) | | Descriptor: | CARBONATE ION, CITRIC ACID, Neutral endopeptidase, ... | | Authors: | Anderson, B.F, Knapp, K.M, Holland, R, Norris, G.E, Christensson, C, Bratt, H, Collins, L.J, Coolbear, T, Lubbers, M.W, Toole, P.W.O, Reid, J.R, Jameson, G.B. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20)
TO BE PUBLISHED
|
|
4J0B
 
 | | Structure of mitochondrial Hsp90 (TRAP1) with ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, COBALT (II) ION, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-30 | | Release date: | 2014-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
3F5J
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
2DLN
 
 | | VANCOMYCIN RESISTANCE: STRUCTURE OF D-ALANINE:D-ALANINE LIGASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 1(S)-AMINOETHYL-(2-CARBOXYPROPYL)PHOSPHORYL-PHOSPHINIC ACID, ADENOSINE-5'-DIPHOSPHATE, D-ALANINE--D-ALANINE LIGASE, ... | | Authors: | Knox, J.R, Moews, P.C, Fan, C. | | Deposit date: | 1994-07-18 | | Release date: | 1995-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Vancomycin resistance: structure of D-alanine:D-alanine ligase at 2.3 A resolution.
Science, 266, 1994
|
|
3FPD
 
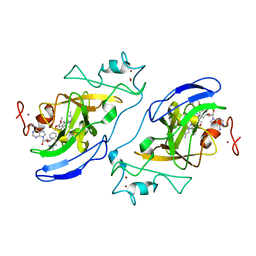 | | G9a-like protein lysine methyltransferase inhibition by BIX-01294 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, N-(1-benzylpiperidin-4-yl)-6,7-dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)quinazolin-4-amine, ... | | Authors: | Chang, Y, Zhang, X, Horton, J.R, Cheng, X. | | Deposit date: | 2009-01-05 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4IVG
 
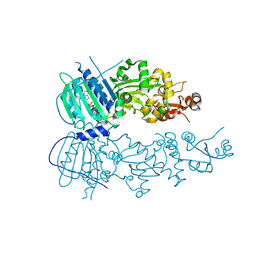 | | Crystal structure of mitochondrial Hsp90 (TRAP1) NTD-Middle domain dimer with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-22 | | Release date: | 2014-01-22 | | Last modified: | 2014-08-27 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
2A9U
 
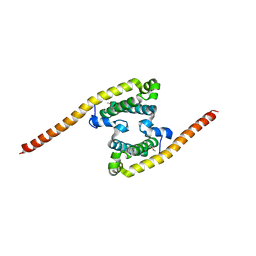 | | Structure of the N-terminal domain of Human Ubiquitin carboxyl-terminal hydrolase 8 (USP8) | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 8 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, E, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-12 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Amino-terminal Dimerization, NRDP1-Rhodanese Interaction, and Inhibited Catalytic Domain Conformation of the Ubiquitin-specific Protease 8 (USP8).
J.Biol.Chem., 281, 2006
|
|
3FL2
 
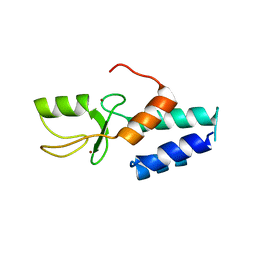 | | Crystal structure of the ring domain of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ring Domain of the E3 Ubiquitin-Protein Ligase Uhrf1
To be Published
|
|
2ABJ
 
 | | Crystal structure of human branched chain amino acid transaminase in a complex with an inhibitor, C16H10N2O4F3SCl, and pyridoxal 5' phosphate. | | Descriptor: | Branched-chain-amino-acid aminotransferase, cytosolic, N'-(5-CHLOROBENZOFURAN-2-CARBONYL)-2-(TRIFLUOROMETHYL)BENZENESULFONOHYDRAZIDE, ... | | Authors: | Ohren, J.F, Moreland, D.W, Rubin, J.R, Hu, H.L, McConnell, P.C, Mistry, A, Mueller, W.T, Scholten, J.D, Hasemann, C.H. | | Deposit date: | 2005-07-15 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The design and synthesis of human branched-chain amino acid aminotransferase inhibitors for treatment of neurodegenerative diseases.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3NV0
 
 | | Crystal structure and mutational analysis of the NXF2/NXT1 heterodimeric complex from caenorhabditis elegans at 1.84 A resolution | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kerkow, D.E, Carmel, A.B, Williamson, J.R. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structure of the NXF2/NXT1 heterodimeric complex reveals the combined specificity and versatility of the NTF2-like fold.
J.Mol.Biol., 415, 2012
|
|
3FL7
 
 | | Crystal structure of the human ephrin A2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Ephrin receptor, ... | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3O8R
 
 | |
3F2E
 
 | | Crystal structure of Yellowstone SIRV coat protein C-terminus | | Descriptor: | CITRIC ACID, SIRV coat protein | | Authors: | Taurog, R.E, Szymczyna, B.R, Williamson, J.R, Johnson, J.E. | | Deposit date: | 2008-10-29 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | Synergy of NMR, computation, and X-ray crystallography for structural biology.
Structure, 17, 2009
|
|
2BC5
 
 | | Crystal structure of E. coli cytochrome b562 with engineered c-type heme linkages | | Descriptor: | HEME C, SULFATE ION, Soluble cytochrome b562 | | Authors: | Faraone-Mennella, J, Tezcan, F.A, Gray, H.B, Winkler, J.R. | | Deposit date: | 2005-10-18 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stability and Folding Kinetics of Structurally Characterized Cytochrome c-b(562).
Biochemistry, 45, 2006
|
|
2DN3
 
 | | 1.25A resolution crystal structure of human hemoglobin in the carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha subunit, Hemoglobin beta subunit, ... | | Authors: | Park, S.-Y, Yokoyama, T, Shibayama, N, Shiro, Y, Tame, J.R. | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 a resolution crystal structures of human haemoglobin in the oxy, deoxy and carbonmonoxy forms.
J.Mol.Biol., 360, 2006
|
|
2DHQ
 
 | |
2DN2
 
 | | 1.25A resolution crystal structure of human hemoglobin in the deoxy form | | Descriptor: | Hemoglobin alpha subunit, Hemoglobin beta subunit, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, S.-Y, Yokoyama, T, Shibayama, N, Shiro, Y, Tame, J.R. | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 a resolution crystal structures of human haemoglobin in the oxy, deoxy and carbonmonoxy forms.
J.Mol.Biol., 360, 2006
|
|
2DSM
 
 | | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450 | | Descriptor: | Hypothetical protein yqaI | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-01 | | Release date: | 2006-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450
to be published
|
|
4N21
 
 | | Crystal structure of the GP2 Core Domain from the California Academy of Science Virus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GP2 Ectodomain | | Authors: | Malashkevich, V.N, Koellhoffer, J.F, Dai, Z, Toro, R, Lai, J.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Characterization of the Glycoprotein GP2 Core Domain from the CAS Virus, a Novel Arenavirus-Like Species.
J.Mol.Biol., 426, 2014
|
|
3TUG
 
 | | Crystal structure of the HECT domain of ITCH E3 ubiquitin ligase | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Itchy homolog, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dobrovetsky, E, Xue, S, Butler, C, Wernimont, A, Walker, J.R, Tempel, W, Dhe-Paganon, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-16 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of the HECT domain of ITCH E3 ubiquitin ligase
To be Published
|
|
4N4Q
 
 | | Crystal Structure of N-acetylneuraminate lyase from Mycoplasma synoviae, crystal form II | | Descriptor: | Acylneuraminate lyase | | Authors: | Georgescauld, F, Popova, K, Gupta, A.J, Bracher, A, Engen, J.R, Hayer-Hartl, M, Hartl, F.U. | | Deposit date: | 2013-10-08 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GroEL/ES Chaperonin Modulates the Mechanism and Accelerates the Rate of TIM-Barrel Domain Folding.
Cell(Cambridge,Mass.), 157, 2014
|
|
4MT0
 
 | |
1PC9
 
 | | Crystal Structure of BnSP-6, a Lys49-Phospholipase A2 | | Descriptor: | BnSP-6 | | Authors: | Magro, A.J, Soares, A.M, Giglio, J.R, Fontes, M.R.M. | | Deposit date: | 2003-05-16 | | Release date: | 2004-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of BnSP-7 and BnSP-6, two Lys49-phospholipases A(2): quaternary structure and inhibition mechanism insights.
Biochem.Biophys.Res.Commun., 311, 2003
|
|
3U3Z
 
 | | Structure of human microcephalin (MCPH1) tandem BRCT domains in complex with an H2A.X peptide phosphorylated at Ser139 and Tyr142 | | Descriptor: | GLYCEROL, Histone H2A.X peptide, Microcephalin | | Authors: | Singh, N, Thompson, J.R, Heroux, A, Mer, G. | | Deposit date: | 2011-10-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dual recognition of phosphoserine and phosphotyrosine in histone variant H2A.X by DNA damage response protein MCPH1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4MZ0
 
 | |
