5CUL
 
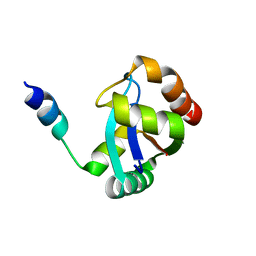 | | crystal structure of the PscU C-terminal domain | | Descriptor: | Translocation protein in type III secretion | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2015-07-24 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of a Type 3 Secretion System (T3SS) Ruler Protein Suggests a Molecular Mechanism for Needle Length Sensing.
J.Biol.Chem., 291, 2016
|
|
4BHU
 
 | | Crystal structure of BslA - A bacterial hydrophobin | | Descriptor: | CHLORIDE ION, GLYCEROL, UNCHARACTERIZED PROTEIN YUAB | | Authors: | Rao, F.V, Hobley, L, Ostrowski, A, Bromley, K.M, Porter, M, Prescott, A.R, Swedlow, J.R, MacPhee, C.E, van Aalten, D.M.F, Stanley-Wall, N.R. | | Deposit date: | 2013-04-08 | | Release date: | 2013-08-14 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Bsla is a Self-Assembling Bacterial Hydrophobin that Coats the Bacillus Subtilis Biofilm.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4YU0
 
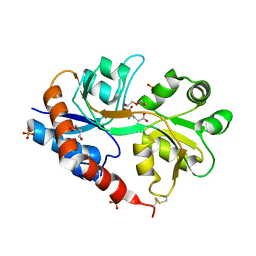 | | Crystal structure of a tetramer of GluA2 TR mutant ligand binding domains bound with glutamate at 1.26 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTAMIC ACID, Glutamate receptor 2,Glutamate receptor 2, ... | | Authors: | Chebli, M, Salazar, H, Baranovic, J, Carbone, A.L, Ghisi, V, Faelber, K, Lau, A.Y, Daumke, O, Plested, A.J.R. | | Deposit date: | 2015-03-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the tetrameric GluA2 ligand-binding domain in complex with glutamate at 1.26 Angstroms resolution
To Be Published
|
|
5CUK
 
 | | Crystal structure of the PscP SS domain | | Descriptor: | GLYCEROL, Ruler protein | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2015-07-24 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of a Type 3 Secretion System (T3SS) Ruler Protein Suggests a Molecular Mechanism for Needle Length Sensing.
J.Biol.Chem., 291, 2016
|
|
1NYG
 
 | |
2ZFY
 
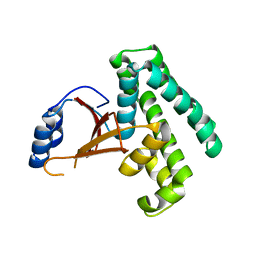 | | Crystal structure of human Otubain 1 | | Descriptor: | Ubiquitin thioesterase OTUB1 | | Authors: | Akutsu, M, Walker, J.R, Li, Y, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis and specificity of human otubain 1-mediated deubiquitination.
Biochem.J., 418, 2009
|
|
4Z0I
 
 | | Crystal structure of a tetramer of GluA2 ligand binding domains bound with glutamate at 1.45 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTAMIC ACID, Glutamate receptor 2,Glutamate receptor 2, ... | | Authors: | Baranovic, J, Chebli, M, Salazar, H, Carbone, A.L, Ghisi, V, Faelber, K, Lau, A.Y, Daumke, O, Plested, A.J.R. | | Deposit date: | 2015-03-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the tetrameric wt GluA2 ligand-binding domain bound to glutamate at 1.45 Angstroms resolution
To Be Published
|
|
4G08
 
 | |
4E36
 
 | | Crystal structure of the human Endoplasmic Reticulum Aminopeptidase 2 variant N392K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Birtley, J.R, Saridakis, E, Pegias, P, Stratikos, E, Mavridis, I.M. | | Deposit date: | 2012-03-09 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A common single nucleotide polymorphism in endoplasmic reticulum aminopeptidase 2 induces a specificity switch that leads to altered antigen processing.
J.Immunol., 189, 2012
|
|
1QKB
 
 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KVK | | Descriptor: | ACETATE ION, PEPTIDE LYS-VAL-LYS, PERIPLASMIC OLIGOPEPTIDE-BINDING PROTEIN, ... | | Authors: | Tame, J.R.H, Sleigh, S.H, Wilkinson, A.J. | | Deposit date: | 1999-07-14 | | Release date: | 1999-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and Calorimetric Analysis of Peptide Binding to Oppa Protein
J.Mol.Biol., 291, 1999
|
|
1QKA
 
 | |
5TIR
 
 | | Crystal Structure of Mn Superoxide Dismutase mutant M27V from Trichoderma reesei | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Mendoza, E.R, Brandao-Neto, J, Pereira, H.M, Ferreira Junior, J.R.S, Garratt, R.C. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of Mn Superoxide Dismutase mutant M27V from Trichoderma reesei
To Be Published
|
|
1RKM
 
 | | STRUCTURE OF OPPA | | Descriptor: | OLIGO-PEPTIDE BINDING PROTEIN | | Authors: | Sleigh, S.H, Tame, J.R.H, Wilkinson, A.J. | | Deposit date: | 1997-03-25 | | Release date: | 1997-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine.
Biochemistry, 36, 1997
|
|
5UGY
 
 | | Influenza hemagglutinin in complex with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH65 heavy chain, ... | | Authors: | Whittle, J.R.R, Jenni, S, Harrison, S.C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin.
Proc. Natl. Acad. Sci. U.S.A., 108, 2011
|
|
6G6P
 
 | | Crystal structure of the computationally designed Ika8 protein: crystal packing No.2 in P63 | | Descriptor: | Ika8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
6G6M
 
 | | Crystal structure of the computationally designed Tako8 protein in P42212 | | Descriptor: | SULFATE ION, Tako8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
6G6Q
 
 | | Crystal structure of the computationally designed Ika4 protein | | Descriptor: | Ika4 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
5TCP
 
 | | Near-atomic resolution cryo-EM structure of the periplasmic domains of PrgH and PrgK | | Descriptor: | Lipoprotein PrgK, Protein PrgH | | Authors: | Worrall, L.J, Hong, C, Vuckovic, M, Bergeron, J.R.C, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2016-09-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body.
Nature, 540, 2016
|
|
6G6N
 
 | | Crystal structure of the computationally designed Tako8 protein in C2 | | Descriptor: | Tako8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
6NU7
 
 | |
6NUM
 
 | |
6NUN
 
 | |
6NOB
 
 | |
6NU8
 
 | |
5TCQ
 
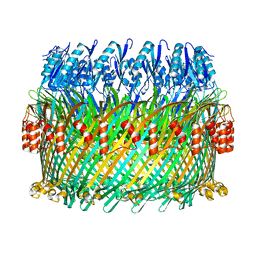 | | Near-atomic resolution cryo-EM structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG | | Descriptor: | Protein InvG | | Authors: | Worrall, L.J, Hong, C, Vuckovic, M, Bergeron, J.R.C, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2016-09-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body.
Nature, 540, 2016
|
|
