2AKJ
 
 | | Structure of spinach nitrite reductase | | Descriptor: | Ferredoxin--nitrite reductase, chloroplast, IRON/SULFUR CLUSTER, ... | | Authors: | Swamy, U, Wang, M, Tripathy, J.N, Kim, S.-K, Hirasawa, M, Knaff, D.B, Allen, J.P. | | Deposit date: | 2005-08-03 | | Release date: | 2006-01-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Spinach Nitrite Reductase: Implications for Multi-electron Reactions by the Iron-Sulfur:Siroheme Cofactor
Biochemistry, 44, 2005
|
|
7P92
 
 | | TmHydABC- T. maritima bifurcating hydrogenase with bridge domain up | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P8N
 
 | | TmHydABC- T. maritima hydrogenase with bridge closed | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P91
 
 | | TmHydABC- T. maritima bifurcating hydrogenase with bridge domain closed | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P5H
 
 | | TmHydABC- D2 map | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-14 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7OVM
 
 | | Protein kinase MKK7 in complex with cyclobutyl-substituted indazole | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7, ~{N}-[(1-cyclobutyl-1,2,3-triazol-4-yl)methyl]-3-(1~{H}-indazol-3-yl)-5-(propanoylamino)benzamide | | Authors: | Buehrmann, M, Wiese, J.N, Mueller, M.P, Rauh, D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Optimization of Covalent MKK7 Inhibitors via Crude Nanomole-Scale Libraries.
J.Med.Chem., 65, 2022
|
|
7OVN
 
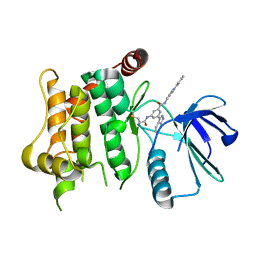 | | Protein kinase MKK7 in complex with tolyl-substituted indazole | | Descriptor: | 3-(1~{H}-indazol-3-yl)-~{N}-[[1-(2-methylphenyl)-1,2,3-triazol-4-yl]methyl]-5-(propanoylamino)benzamide, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Buehrmann, M, Wiese, J.N, Mueller, M.P, Rauh, D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Optimization of Covalent MKK7 Inhibitors via Crude Nanomole-Scale Libraries.
J.Med.Chem., 65, 2022
|
|
7OVJ
 
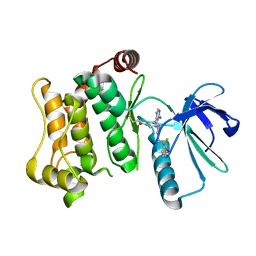 | | Protein kinase MKK7 in complex with difluoro-phenethyltriazole-substituted pyrazolopyrimidine | | Descriptor: | 1-[(3~{R})-3-[4-azanyl-3-[1-[2,2-bis(fluoranyl)-2-phenyl-ethyl]-1,2,3-triazol-4-yl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]propan-1-one, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of Covalent MKK7 Inhibitors via Crude Nanomole-Scale Libraries.
J.Med.Chem., 65, 2022
|
|
7OVL
 
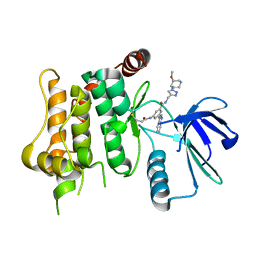 | | Protein kinase MKK7 in complex with methoxycyclohexyl-substituted indazole | | Descriptor: | 3-(2~{H}-indazol-3-yl)-~{N}-[[1-[(1~{R},2~{R})-2-methoxycyclohexyl]-1,2,3-triazol-4-yl]methyl]-5-(propanoylamino)benzamide, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Buehrmann, M, Wiese, J.N, Mueller, M.P, Rauh, D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Optimization of Covalent MKK7 Inhibitors via Crude Nanomole-Scale Libraries.
J.Med.Chem., 65, 2022
|
|
7P4R
 
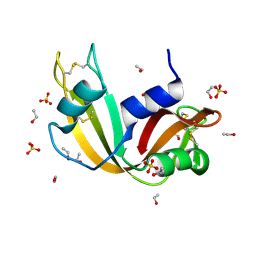 | | Ultra High Resolution X-ray Structure of Orthorhombic Bovine Pancreatic Ribonuclease at 100K | | Descriptor: | ETHANOL, Ribonuclease pancreatic, SULFATE ION | | Authors: | Lisgarten, D.R, Palmer, R.A, Cooper, J.B, Naylor, C.E, Howlin, B.J, Lisgarten, J.N, Najmudin, S, Lobley, C.M.C. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ultra-high resolution X-ray structure of orthorhombic bovine pancreatic Ribonuclease A at 100K.
BMC Chem, 17, 2023
|
|
1OPF
 
 | |
4NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
3TPZ
 
 | |
2GSA
 
 | |
1PII
 
 | |
1TAS
 
 | |
2DKB
 
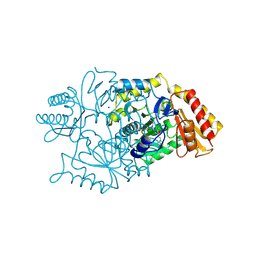 | | DIALKYLGLYCINE DECARBOXYLASE STRUCTURE: BIFUNCTIONAL ACTIVE SITE AND ALKALI METAL BINDING SITES | | Descriptor: | 2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Toney, M.D, Hohenester, E, Jansonius, J.N. | | Deposit date: | 1994-07-12 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dialkylglycine decarboxylase structure: bifunctional active site and alkali metal sites.
Science, 261, 1993
|
|
3L1T
 
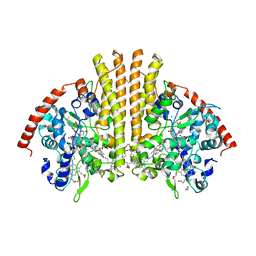 | | E. coli NrfA sulfite ocmplex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Hemmings, A.M, Butt, J.N. | | Deposit date: | 2009-12-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and thermodynamic resolution of the interactions between sulfite and the pentahaem cytochrome NrfA from Escherichia coli
Biochem.J., 431, 2010
|
|
2QNY
 
 | | Crystal structure of the complex between the A246F mutant of mycobacterium beta-ketoacyl-acyl carrier protein synthase III (FABH) and SS-(2-hydroxyethyl) O-decyl ester carbono(dithioperoxoic) acid | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, BETA-MERCAPTOETHANOL, DECYL FORMATE | | Authors: | Sachdeva, S, Musayev, F, Alhamadsheh, M, Scarsdale, J.N, Wright, H.T, Reynolds, K.A. | | Deposit date: | 2007-07-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Separate Entrance and Exit Portals for Ligand Traffic in Mycobacterium tuberculosis FabH
Chem.Biol., 15, 2008
|
|
3K0H
 
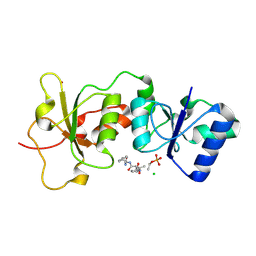 | | The crystal structure of BRCA1 BRCT in complex with a minimal recognition tetrapeptide with an amidated C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3K15
 
 | | Crystal Structure of BRCA1 BRCT D1840T in complex with a minimal recognition tetrapeptide with an amidated C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-25 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3K16
 
 | | Crystal Structure of BRCA1 BRCT D1840T in complex with a minimal recognition tetrapeptide with a free carboxy C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-25 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
2QNZ
 
 | | Crystal structure of the complex between the mycobacterium beta-ketoacyl-acyl carrier protein synthase III (FABH) and SS-(2-hydroxyethyl)-O-decyl ester carbono(dithioperoxoic) acid | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, BETA-MERCAPTOETHANOL, DECYL FORMATE | | Authors: | Sachdeva, S, Musayev, F, Alhamadsheh, M, Scarsdale, J.N, Wright, H.T, Reynolds, K.A. | | Deposit date: | 2007-07-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Separate Entrance and Exit Portals for Ligand Traffic in Mycobacterium tuberculosis FabH
Chem.Biol., 15, 2008
|
|
3K05
 
 | |
3K0K
 
 | | Crystal Structure of BRCA1 BRCT in complex with a minimal recognition tetrapeptide with a free carboxy C-terminus. | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
