5ES7
 
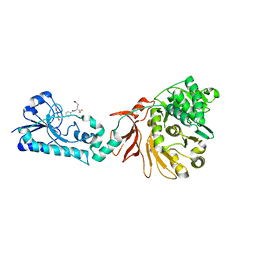 | | Crystal structure of the F-A domains of the LgrA initiation module soaked with FON, AMPcPP, and valine. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Linear gramicidin synthetase subunit A, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Reimer, J.M, Aloise, M.N, Schmeing, T.M. | | Deposit date: | 2015-11-16 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Synthetic cycle of the initiation module of a formylating nonribosomal peptide synthetase.
Nature, 529, 2016
|
|
5EUC
 
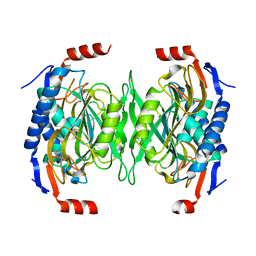 | | The role of the C-terminal region on the oligomeric state and enzymatic activity of Trypanosoma cruzi hypoxanthine phosphoribosyl transferase | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Valsecchi, W.M, Cousido-Siah, A, Mitschler, A, Podjarny, A, Delfino, J.M, Santos, J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The role of the C-terminal region on the oligomeric state and enzymatic activity of Trypanosoma cruzi hypoxanthine phosphoribosyl transferase.
Biochim.Biophys.Acta, 1864, 2016
|
|
6OEH
 
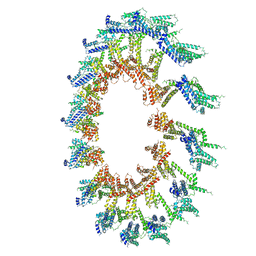 | | PolyAla Model of OMCC I-Layer | | Descriptor: | PolyAla Model of OMCC I-Layer | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
5ESD
 
 | | Crystal Structure of M. tuberculosis MenD bound to ThDP and Mn2+ | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, MANGANESE (II) ION, THIAMINE DIPHOSPHATE | | Authors: | Johnston, J.M, Jirgis, E.N.M, Bashiri, G, Bulloch, E.M.M, Baker, E.N. | | Deposit date: | 2015-11-16 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Views along the Mycobacterium tuberculosis MenD Reaction Pathway Illuminate Key Aspects of Thiamin Diphosphate-Dependent Enzyme Mechanisms.
Structure, 24, 2016
|
|
5DBT
 
 | |
6OM5
 
 | | Structure of a haemophore from Haemophilus haemolyticus | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Torrado, M, Walshe, J.L, Mackay, J.P, Guss, J.M, Gell, D.A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A heme-binding protein produced by Haemophilus haemolyticus inhibits non-typeable Haemophilus influenzae.
Mol.Microbiol., 113, 2020
|
|
6OMY
 
 | | Protein Tyrosine Phosphatase 1B (1-301), P180A mutant, apo state | | Descriptor: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Cui, D.S, Lipchock, J.M, Loria, J.P. | | Deposit date: | 2019-04-19 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
5HBS
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with all-trans-retinol at 0.89 angstrom. | | Descriptor: | RETINOL, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2016-01-02 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
6O9T
 
 | | KirBac3.1 mutant at a resolution of 4.1 Angstroms | | Descriptor: | 2,3,5,6-tetramethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, Inward rectifier potassium channel Kirbac3.1, POTASSIUM ION | | Authors: | Gulbis, J.M, Black, K.A, Miller, D.M. | | Deposit date: | 2019-03-15 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
6O9V
 
 | | KirBac3.1 mutant at a resolution of 3.1 Angstroms | | Descriptor: | 1,1-Methanediyl Bismethanethiosulfonate, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, Inward rectifier potassium channel Kirbac3.1, ... | | Authors: | Gulbis, J.M, Black, K.A, Miller, D.M. | | Deposit date: | 2019-03-15 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.094 Å) | | Cite: | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
2X7L
 
 | | Implications of the HIV-1 Rev dimer structure at 3.2A resolution for multimeric binding to the Rev response element | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, PROTEIN REV | | Authors: | DiMattia, M.A, Watts, N.R, Stahl, S.J, Rader, C, Wingfield, P.T, Stuart, D.I, Steven, A.C, Grimes, J.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Implications of the HIV-1 Rev Dimer Structure at 3. 2 A Resolution for Multimeric Binding to the Rev Response Element.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6OEF
 
 | | PolyAla Model of the O-layer from the Type 4 Secretion System of H. pylori | | Descriptor: | PolyAla Model of OMCC O-Layer | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
1HHT
 
 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 plus template | | Descriptor: | DNA (5'-(*TP*TP*TP*CP*C)-3'), MANGANESE (II) ION, P2 PROTEIN | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2000-12-28 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
5HA1
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with retinylamine | | Descriptor: | (2~{E},4~{E},6~{E},8~{E})-3,7-dimethyl-9-(2,6,6-trimethylcyclohexen-1-yl)nona-2,4,6,8-tetraen-1-amine, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
6OW4
 
 | | Structure of the NADH-bound form of 20beta-Hydroxysteroid Dehydrogenase from Bifidobacterium adolescentis strain L2-32 | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase, short chain dehydrogenase/reductase family protein | | Authors: | Mythen, S.M, Pollet, R.M, Koropatkin, N.M, Ridlon, J.M. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and biochemical characterization of 20 beta-hydroxysteroid dehydrogenase fromBifidobacterium adolescentisstrain L2-32.
J.Biol.Chem., 294, 2019
|
|
1HHS
 
 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 | | Descriptor: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2000-12-28 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
5DVX
 
 | | Crystal structure of the catalytic-domain of human carbonic anhydrase IX at 1.6 angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Carbonic anhydrase 9, ... | | Authors: | Mahon, B.P, Socorro, L, Driscoll, J.M, McKenna, R. | | Deposit date: | 2015-09-21 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | The Structure of Carbonic Anhydrase IX Is Adapted for Low-pH Catalysis.
Biochemistry, 55, 2016
|
|
5FVC
 
 | | Structure of RNA-bound decameric HMPV nucleoprotein | | Descriptor: | HMPV NUCLEOPROTEIN, RNA | | Authors: | Renner, M, Bertinelli, M, Leyrat, C, Paesen, G.C, Saraiva de Oliveira, L.F, Huiskonen, J.T, Grimes, J.M. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Nucleocapsid assembly in pneumoviruses is regulated by conformational switching of the N protein.
Elife, 5, 2016
|
|
1HLC
 
 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN DIMERIC S-LAC LECTIN, L-14-II, IN COMPLEX WITH LACTOSE AT 2.9 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN LECTIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Lobsanov, Y.D, Gitt, M.A, Leffler, H, Barondes, S, Rini, J.M. | | Deposit date: | 1993-10-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystal structure of the human dimeric S-Lac lectin, L-14-II, in complex with lactose at 2.9-A resolution.
J.Biol.Chem., 268, 1993
|
|
6OD2
 
 | | Yeast Spc42 C-terminal Antiparallel Coiled-Coil | | Descriptor: | Spindle pole body component SPC42 | | Authors: | Drennan, A.C, Krishna, S, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jaspersen, S.L, Rayment, I. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-24 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
6ODI
 
 | | Structure of CagY from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagY | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
5FEP
 
 | | HydE from T. maritima in complex with (2R,4R)-MeTDA | | Descriptor: | (2R,4R)-2-methyl-1,3-thiazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
5FQJ
 
 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase bound to 5(6)-nitrobenzotriazole (TS-analog) | | Descriptor: | 6-NITROBENZOTRIAZOLE, GNCA LACTAMASE W229D | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
2XU3
 
 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-1-[1-(5-chlorothiophen-2-yl)cyclopropyl]carbonyl-N-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CATHEPSIN L1, ... | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2010-10-14 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Systematic Investigation of Halogen Bonding in Protein-Ligand Interactions.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
5HE9
 
 | |
