1D6I
 
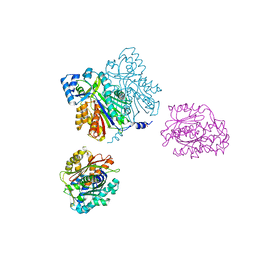 | | CHALCONE SYNTHASE (H303Q MUTANT) | | Descriptor: | CHALCONE SYNTHASE, SULFATE ION | | Authors: | Jez, J.M, Ferrer, J.L, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 1999-10-13 | | Release date: | 2000-02-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase.
Biochemistry, 39, 2000
|
|
1CKQ
 
 | |
1CL8
 
 | |
7SX1
 
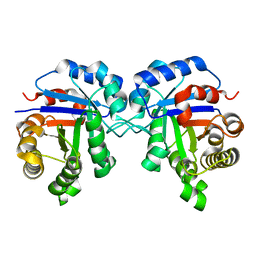 | | human triosephosphate isomerase mutant v154m | | Descriptor: | ISOPROPYL ALCOHOL, Triosephosphate isomerase | | Authors: | Romero, J.M. | | Deposit date: | 2021-11-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | human triosephosphate isomerase mutant v154m
To Be Published
|
|
7T0Q
 
 | | human triosephosphate isomerase mutant v154m | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Romero, J.M. | | Deposit date: | 2021-11-30 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | human triosephosphate isomerase mutant v154m
To Be Published
|
|
1D3B
 
 | | CRYSTAL STRUCTURE OF THE D3B SUBCOMPLEX OF THE HUMAN CORE SNRNP DOMAIN AT 2.0A RESOLUTION | | Descriptor: | CITRIC ACID, GLYCEROL, PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN ASSOCIATED PROTEIN B), ... | | Authors: | Kambach, C, Walke, S, Avis, J.M, De La Fortelle, E, Li, J, Nagai, K. | | Deposit date: | 1998-12-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs.
Cell(Cambridge,Mass.), 96, 1999
|
|
1D3Y
 
 | | STRUCTURE OF THE DNA TOPOISOMERASE VI A SUBUNIT | | Descriptor: | DNA TOPOISOMERASE VI A SUBUNIT, MAGNESIUM ION, SODIUM ION | | Authors: | Nichols, M.D, DeAngelis, K.A, Keck, J.L, Berger, J.M. | | Deposit date: | 1999-10-01 | | Release date: | 1999-11-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11.
EMBO J., 18, 1999
|
|
1D5F
 
 | | STRUCTURE OF E6AP: INSIGHTS INTO UBIQUITINATION PATHWAY | | Descriptor: | E6AP HECT CATALYTIC DOMAIN, E3 LIGASE | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-07 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
7TFM
 
 | | Atomic Structure of the Leishmania spp. Hsp100 N-Domain | | Descriptor: | ATP-dependent Clp protease subunit, heat shock protein 100 (HSP100), GLYCEROL | | Authors: | Mercado, J.M, Lee, S, Chang, C, Sung, N, Soong, L, Catic, A, Tsai, F.T.F. | | Deposit date: | 2022-01-06 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.055 Å) | | Cite: | Atomic structure of the Leishmania spp. Hsp100 N-domain.
Proteins, 90, 2022
|
|
1D6F
 
 | | CHALCONE SYNTHASE C164A MUTANT | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHALCONE SYNTHASE, SULFATE ION | | Authors: | Jez, J.M, Ferrer, J.L, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 1999-10-13 | | Release date: | 2000-02-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase.
Biochemistry, 39, 2000
|
|
5N2O
 
 | | Structure Of P63 SAM Domain L514F Mutant Causative Of AEC Syndrome | | Descriptor: | Tumor protein 63 | | Authors: | Rinnenthal, J, Wuerz, J.M, Osterburg, C, Guentert, P, Doetsch, V. | | Deposit date: | 2017-02-08 | | Release date: | 2018-02-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein aggregation of the p63 transcription factor underlies severe skin fragility in AEC syndrome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1D6H
 
 | | CHALONE SYNTHASE (N336A MUTANT COMPLEXED WITH COA) | | Descriptor: | CHALCONE SYNTHASE, COENZYME A, SULFATE ION | | Authors: | Jez, J.M, Ferrer, J.L, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 1999-10-13 | | Release date: | 2000-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase.
Biochemistry, 39, 2000
|
|
1DB2
 
 | | CRYSTAL STRUCTURE OF NATIVE PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Nar, H, Bauer, M, Stassen, J.M, Lang, D, Gils, A, Declerck, P. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasminogen activator inhibitor 1. Structure of the native serpin, comparison to its other conformers and implications for serpin inactivation.
J.Mol.Biol., 297, 2000
|
|
7TSR
 
 | | Room temperature rsEospa Cis-state structure at pH 8.4 | | Descriptor: | Cis-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
1DCV
 
 | |
5QDC
 
 | | Crystal structure of BACE complex with BMC019 hydrolyzed | | Descriptor: | (4S)-4-[(1R)-1,2-dihydroxyethyl]-N,N-dimethyl-2-oxo-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaene-19-carboxamide, Beta-secretase 1, GLYCEROL | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1DDE
 
 | | STRUCTURE OF THE DNAG CATALYTIC CORE | | Descriptor: | DNA PRIMASE, YTTRIUM ION | | Authors: | Keck, J.L, Roche, D.D, Lynch, A.S, Berger, J.M. | | Deposit date: | 1999-11-09 | | Release date: | 2000-04-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the RNA polymerase domain of E. coli primase.
Science, 287, 2000
|
|
1DHR
 
 | | CRYSTAL STRUCTURE OF RAT LIVER DIHYDROPTERIDINE REDUCTASE | | Descriptor: | DIHYDROPTERIDINE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Varughese, K.I, Skinner, M.M, Whiteley, J.M, Matthews, D.A, Xuong, N.H. | | Deposit date: | 1992-03-30 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of rat liver dihydropteridine reductase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
7TSS
 
 | | Room temperature rsEospa Trans-state structure at pH 8.4 | | Descriptor: | Trans-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
7TSU
 
 | | Room temperature rsEospa Cis-state structure at pH 5.5 | | Descriptor: | Cis-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
1DI1
 
 | | CRYSTAL STRUCTURE OF ARISTOLOCHENE SYNTHASE FROM PENICILLIUM ROQUEFORTI | | Descriptor: | ARISTOLOCHENE SYNTHASE | | Authors: | Caruthers, J.M, Kang, I, Cane, D.E, Christianson, D.W, Rynkiewicz, M.J. | | Deposit date: | 1999-11-28 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure determination of aristolochene synthase from the blue cheese mold, Penicillium roqueforti.
J.Biol.Chem., 275, 2000
|
|
1DKF
 
 | | CRYSTAL STRUCTURE OF A HETERODIMERIC COMPLEX OF RAR AND RXR LIGAND-BINDING DOMAINS | | Descriptor: | 4-[(4,4-DIMETHYL-1,2,3,4-TETRAHYDRO-[1,2']BINAPTHALENYL-7-CARBONYL)-AMINO]-BENZOIC ACID, OLEIC ACID, PROTEIN (RETINOIC ACID RECEPTOR-ALPHA), ... | | Authors: | Bourguet, W, Vivat, V, Wurtz, J.M, Chambon, P, Gronemeyer, H, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1999-12-07 | | Release date: | 2000-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains.
Mol.Cell, 5, 2000
|
|
1DDK
 
 | | CRYSTAL STRUCTURE OF IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, ZINC ION | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
7TSV
 
 | | Room temperature rsEospa Trans-state structure at pH 5.5 | | Descriptor: | Trans-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
1DFP
 
 | | FACTOR D INHIBITED BY DIISOPROPYL FLUOROPHOSPHATE | | Descriptor: | DIISOPROPYL PHOSPHONATE, FACTOR D | | Authors: | Cole, L.B, Chu, N, Kilpatrick, J.M, Volanakis, J.E, Narayana, S.V.L, Babu, Y.S. | | Deposit date: | 1997-02-18 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of diisopropyl fluorophosphate-inhibited factor D.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
