4IM8
 
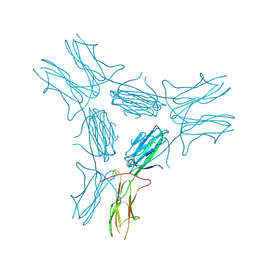 | | low resolution crystal structure of mouse RAGE | | Descriptor: | Advanced glycation end-products receptor | | Authors: | Xu, D, Young, J.H, Krahn, J.M, Song, D, Corbett, K.D, Chazin, W.J, Pedersen, L.C, Esko, J.D. | | Deposit date: | 2013-01-02 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Stable RAGE-Heparan Sulfate Complexes Are Essential for Signal Transduction.
Acs Chem.Biol., 8, 2013
|
|
4JXW
 
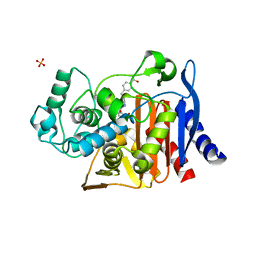 | |
4JO8
 
 | | Crystal structure of the activating Ly49H receptor in complex with m157 (G1F strain) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor 8, M157 | | Authors: | Berry, R, Ng, N, Saunders, P.M, Vivian, J.P, Lin, J, Deuss, F.A, Corbett, A.J, Forbes, C.A, Widjaja, J.M, Sullivan, L.C, McAlister, A.D, Perugini, M.A, Call, M.J, Scalzo, A.A, Degli-Esposti, M.A, Coudert, J.D, Beddoe, T, Brooks, A.G, Rossjohn, J. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Targeting of a natural killer cell receptor family by a viral immunoevasin
Nat.Immunol., 14, 2013
|
|
8UPT
 
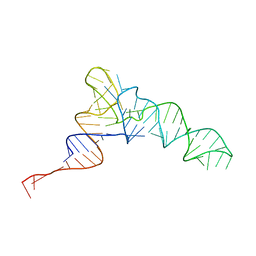 | | Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome | | Descriptor: | RNA (71-MER) | | Authors: | Krahn, N, Zhang, J, Melnikov, S.V, Tharp, J.M, Villa, A, Patel, A, Howard, R.J, Gabir, H, Patel, T.R, Stetefeld, J, Puglisi, J, Soll, D. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | tRNA shape is an identity element for an archaeal pyrrolysyl-tRNA synthetase from the human gut.
Nucleic Acids Res., 52, 2024
|
|
4J9Z
 
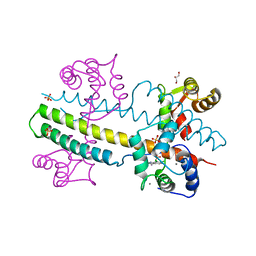 | | Calcium-calmodulin complexed with the calmodulin binding domain from a small conductance potassium channel splice variant and NS309 | | Descriptor: | (3E)-6,7-dichloro-3-(hydroxyimino)-1,3-dihydro-2H-indol-2-one, CALCIUM ION, Calmodulin, ... | | Authors: | Zhang, M, Pascal, J.M, Zhang, J.-F. | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unstructured to structured transition of an intrinsically disordered protein peptide in coupling Ca2+-sensing and SK channel activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8U8Y
 
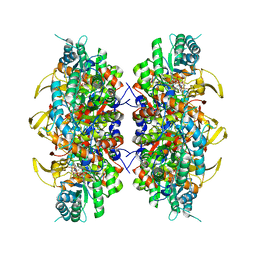 | | Human retinal variant phosphomimetic IMPDH1(546)-S477D filament bound by ATP, IMP, and NAD+, interface-centered | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
8U18
 
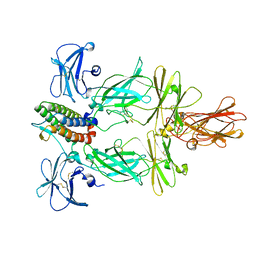 | | Cryo-EM structure of murine Thrombopoietin receptor ectodomain in complex with Tpo | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombopoietin, Thrombopoietin receptor,GCN4 isoform 1, ... | | Authors: | Sarson-Lawrence, K.S, Hardy, J.M, Leis, A, Babon, J.J, Kershaw, N.J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the extracellular domain of murine Thrombopoietin Receptor in complex with Thrombopoietin.
Nat Commun, 15, 2024
|
|
8U8O
 
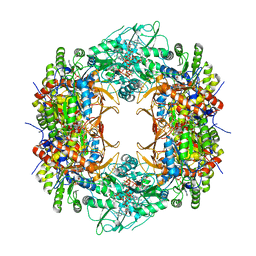 | | Human retinal variant phosphomimetic IMPDH1(546)-S477D filament bound by ATP, IMP, and NAD+, octamer-centered | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 1, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
8U7Q
 
 | | Human retinal variant phosphomimetic IMPDH1(546)-S477D filament bound by GTP, ATP, IMP, and NAD+, octamer-centered | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
4JZQ
 
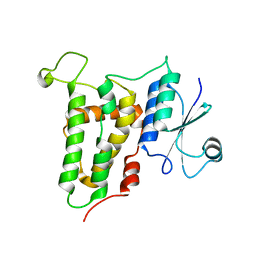 | |
4K0N
 
 | | Crystal structure of human CLIC1 C24A mutant | | Descriptor: | Chloride intracellular channel protein 1, MALONATE ION | | Authors: | Phang, J.M, Harrop, S.J, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2013-04-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure analysis of CLIC1 C24 mutants
To be Published
|
|
4K8I
 
 | | Crystal Structure of Staphylococcal Nuclease mutant I92V/V99L | | Descriptor: | Thermonuclease | | Authors: | Sanders, J.M, Latimer, E.C, Roeser, J.R, Janowska, K, Sakon, J, Stites, W.E. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hydrophobic core mutants of Staphylococcal nuclease
To be Published
|
|
4KSE
 
 | | Crystal structure of a HIV p51 (219-230) deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, HIV p51 subunit | | Authors: | Zheng, X, Mueller, G.A, Derose, E.F, Pedersen, L.C, Gabel, S.A, Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2013-05-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation.
Nucleic Acids Res., 42, 2014
|
|
3L28
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain K339A mutant | | Descriptor: | CHLORIDE ION, Polymerase cofactor VP35, SODIUM ION, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4KRI
 
 | |
4K2F
 
 | | Structure of Pseudomonas aeruginosa PvdQ bound to BRD-A08522488 | | Descriptor: | (2S)-(4-chlorophenyl)(6-chloropyridin-2-yl)ethanenitrile, 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ | | Authors: | Drake, E.J, Wurst, J.M, Theriault, J.R, Munoz, B, Gulick, A.M. | | Deposit date: | 2013-04-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Identification of Inhibitors of PvdQ, an Enzyme Involved in the Synthesis of the Siderophore Pyoverdine.
Acs Chem.Biol., 9, 2014
|
|
4GQB
 
 | | Crystal Structure of the human PRMT5:MEP50 Complex | | Descriptor: | (2S,5S,6E)-2,5-diamino-6-[(3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxydihydrofuran-2(3H)-ylidene]hexanoic acid, Histone H4 peptide, Methylosome protein 50, ... | | Authors: | Antonysamy, S, Bonday, Z, Campbell, R, Doyle, B, Druzina, Z, Gheyi, T, Han, B, Jungheim, L.N, Qian, Y, Rauch, C, Russell, M, Sauder, J.M, Wasserman, S.R, Weichert, K, Willard, F.S, Zhang, A, Emtage, S. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the human PRMT5:MEP50 complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HHE
 
 | | Quinolinate synthase from Pyrococcus furiosus | | Descriptor: | CHLORIDE ION, Quinolinate synthase A | | Authors: | Soriano, E.V, Zhang, Y, Settembre, E.C, Colabroy, K, Sanders, J.M, Dorrestein, P.C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-10-09 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Active-site models for complexes of quinolinate synthase with substrates and intermediates.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H7B
 
 | |
4GW5
 
 | | cQYN meditope - Cetuximab Fab | | Descriptor: | Fab Heavy Chain, Fab Light Chain, PHOSPHATE ION, ... | | Authors: | Donaldson, J.M, Zer, C, Avery, K.N, Bzymek, K.P, Horne, D.A, Williams, J.C. | | Deposit date: | 2012-08-31 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification and grafting of a unique peptide-binding site in the Fab framework of monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KRG
 
 | |
4KRH
 
 | |
4HEA
 
 | | Crystal structure of the entire respiratory complex I from Thermus thermophilus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Baradaran, R, Berrisford, J.M, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3027 Å) | | Cite: | Crystal structure of the entire respiratory complex I.
Nature, 494, 2013
|
|
4GW1
 
 | | cQFD Meditope | | Descriptor: | Fab heavy chain, Fab light chain, PHOSPHATE ION, ... | | Authors: | Donaldson, J.M, Zer, C, Avery, K.N, Bzymek, K.P, Horne, D.A, Williams, J.C. | | Deposit date: | 2012-08-31 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Identification and grafting of a unique peptide-binding site in the Fab framework of monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3L25
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
