3E4C
 
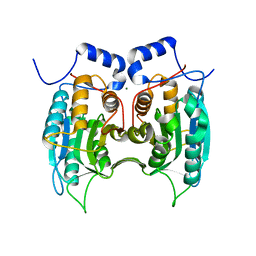 | | Procaspase-1 zymogen domain crystal structure | | Descriptor: | Caspase-1, MAGNESIUM ION | | Authors: | Elliott, J.M, Rouge, L, Wiesmann, C, Scheer, J.M. | | Deposit date: | 2008-08-11 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of procaspase-1 zymogen domain reveals insight into inflammatory caspase autoactivation
J.Biol.Chem., 284, 2009
|
|
1HIN
 
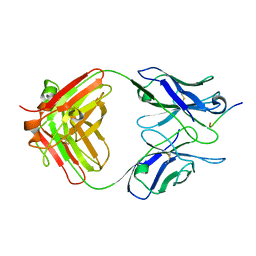 | |
1HI1
 
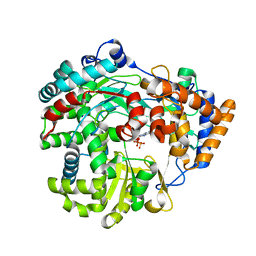 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 plus bound NTP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, P2 PROTEIN | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2001-01-01 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
3G7C
 
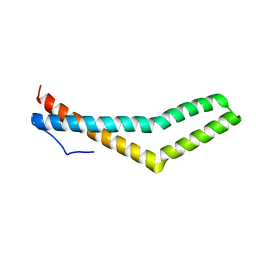 | | Structure of the Phosphorylation Mimetic of Occludin C-term Tail | | Descriptor: | Occludin | | Authors: | Tash, B.R, Sundstrom, J.M, Murakami, T, Flanagan, J.M, Bewley, M.C, Antonetii, D.A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and analysis of occludin phosphosites: a combined mass spectrometry and bioinformatics approach.
J.PROTEOME RES., 8, 2009
|
|
1HI8
 
 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 | | Descriptor: | MAGNESIUM ION, P2 PROTEIN | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2001-01-03 | | Release date: | 2001-03-27 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
3VEK
 
 | | Both Zn Fingers of GATA1 Bound to Palindromic DNA Recognition Site, P1 Crystal Form | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*GP*TP*CP*CP*AP*TP*CP*TP*GP*AP*TP*AP*AP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*TP*TP*AP*TP*CP*AP*GP*AP*TP*GP*GP*AP*CP*TP*C)-3'), Erythroid transcription factor, ... | | Authors: | Jacques, D.A, Ripin, N, Wilkinson-White, L.E, Guss, J.M, Matthews, J.M. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA.
Protein Sci., 24, 2015
|
|
3PI5
 
 | | Crystal Structure of Human Beta Secretase in Complex with BFG356 | | Descriptor: | (3S,4S,5R)-3-(3-bromo-4-hydroxybenzyl)-5-[(3-cyclopropylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1 | | Authors: | Rondeau, J.M. | | Deposit date: | 2010-11-05 | | Release date: | 2011-03-23 | | Last modified: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2G2U
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
2GDQ
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution | | Descriptor: | yitF | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-16 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution
To be Published
|
|
1ZEG
 
 | | STRUCTURE OF B28 ASP INSULIN IN COMPLEX WITH PHENOL | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-05-01 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
3HJ6
 
 | | Structure of Halothermothrix orenii fructokinase (FRK) | | Descriptor: | Fructokinase | | Authors: | Chua, T.K, Seetharaman, J, Kasprzak, J.M, Ng, C, Patel, B.K, Love, C, Bujnicki, J.M, Sivaraman, J. | | Deposit date: | 2009-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a fructokinase homolog from Halothermothrix orenii
J.Struct.Biol., 171, 2010
|
|
5E3J
 
 | | The response regulator RstA is a potential drug target for Acinetobacter baumannii | | Descriptor: | Response regulator RstA | | Authors: | Russo, T.A, Manohar, A, Beanan, J.M, Olson, R, MacDonald, U, Graham, J, Umland, T.C. | | Deposit date: | 2015-10-02 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Response Regulator BfmR Is a Potential Drug Target for Acinetobacter baumannii.
Msphere, 1, 2016
|
|
5ES5
 
 | |
1AX8
 
 | | Human obesity protein, leptin | | Descriptor: | OBESITY PROTEIN | | Authors: | Zhang, F, Beals, J.M, Briggs, S.L, Clawson, D.K, Wery, J.-P, Schevitz, R.W. | | Deposit date: | 1997-10-31 | | Release date: | 1998-11-25 | | Last modified: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the obese protein leptin-E100.
Nature, 387, 1997
|
|
5ES7
 
 | | Crystal structure of the F-A domains of the LgrA initiation module soaked with FON, AMPcPP, and valine. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Linear gramicidin synthetase subunit A, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Reimer, J.M, Aloise, M.N, Schmeing, T.M. | | Deposit date: | 2015-11-16 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Synthetic cycle of the initiation module of a formylating nonribosomal peptide synthetase.
Nature, 529, 2016
|
|
5EZR
 
 | | Crystal Structure of PVX_084705 bound to compound | | Descriptor: | CHLORIDE ION, N-[5-(3-{2-[(cyclopropylmethyl)amino]pyrimidin-4-yl}-7-[(dimethylamino)methyl]-6-methylimidazo[1,2-a]pyridin-2-yl)-2-fluorophenyl]methanesulfonamide, cGMP-dependent protein kinase, ... | | Authors: | El Bakkouri, M, Amani, M, Walker, J.R, Osborne, S, Large, J.M, Birchall, K, Bouloc, N, Smiljanic-Hurley, E, Wheldon, M, Harding, D.J, Merritt, A.T, Ansell, K.H, Coombs, P.J, Kettleborough, C.A, Stewart, B.L, Bowyer, P.W, Gutteridge, W.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Baker, D.A, Hui, R, Loppnau, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-26 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PVX_084705 bound to compound
To Be Published
|
|
5F2E
 
 | | Crystal Structure of small molecule ARS-853 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-[4-[2-[[4-chloranyl-5-(1-methylcyclopropyl)-2-oxidanyl-phenyl]amino]ethanoyl]piperazin-1-yl]azetidin-1-yl]prop-2-en-1-one, GLYCEROL, GLYCINE, ... | | Authors: | Patricelli, M.P, Janes, M.R, Li, L.-S, Hansen, R, Peters, U, Kessler, L.V, Chen, Y, Kucharski, J.M, Feng, J, Ely, T, Chen, J.H, Firdaus, S.J, Babbar, A, Ren, P, Liu, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State.
Cancer Discov, 6, 2016
|
|
5FEX
 
 | | HydE from T. maritima in complex with Se-adenosyl-L-selenocysteine (tfinal of the reaction) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
5FVC
 
 | | Structure of RNA-bound decameric HMPV nucleoprotein | | Descriptor: | HMPV NUCLEOPROTEIN, RNA | | Authors: | Renner, M, Bertinelli, M, Leyrat, C, Paesen, G.C, Saraiva de Oliveira, L.F, Huiskonen, J.T, Grimes, J.M. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Nucleocapsid assembly in pneumoviruses is regulated by conformational switching of the N protein.
Elife, 5, 2016
|
|
5FEP
 
 | | HydE from T. maritima in complex with (2R,4R)-MeTDA | | Descriptor: | (2R,4R)-2-methyl-1,3-thiazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
5FQJ
 
 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase bound to 5(6)-nitrobenzotriazole (TS-analog) | | Descriptor: | 6-NITROBENZOTRIAZOLE, GNCA LACTAMASE W229D | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
1BTL
 
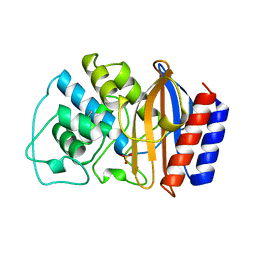 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI TEM1 BETA-LACTAMASE AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | BETA-LACTAMASE TEM1, SULFATE ION | | Authors: | Jelsch, C, Mourey, L, Masson, J.M, Samama, J.P. | | Deposit date: | 1993-11-01 | | Release date: | 1995-01-26 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Escherichia coli TEM1 beta-lactamase at 1.8 A resolution.
Proteins, 16, 1993
|
|
2ND3
 
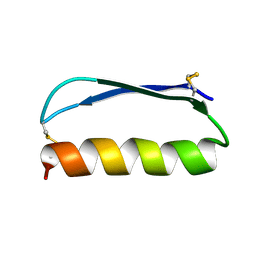 | | Solution structure of the de novo mini protein gEEH_04 | | Descriptor: | De novo mini protein EEH_04 | | Authors: | Pulavarti, S.V, Bahl, C.D, Gilmore, J.M, Eletsky, A, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
5FQQ
 
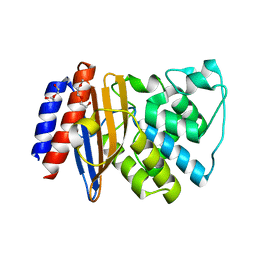 | | Last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, GNCA4 LACTAMASE | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5FXC
 
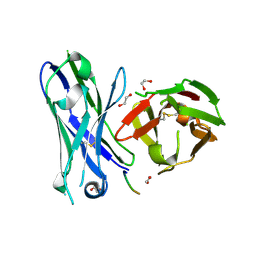 | | Crystal structure of glycopeptide 22 in complex with scFv-SM3 | | Descriptor: | 1,2-ETHANEDIOL, GLYCOPEPTIDE, SCFV-SM3 | | Authors: | Rojas-Ocariz, V, Companon, I, Aydillo, C, Castro-Lopez, J, Jimenez-Barbero, J, Hurtado-Guerrero, R, Avenoza, A, Zurbano, M.M, Corzana, F, Busto, J.H, Peregrina, J.M. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design of Alpha-S-Glycopeptides Derived from Muc1 with a Flexible and Solvent Exposed Sugar Moiety
J.Org.Chem., 81, 2016
|
|
