7RP9
 
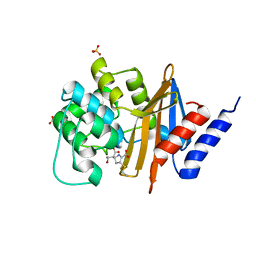 | | X-ray crystal structure of OXA-24/40 V130D in complex with imipenem | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, BICARBONATE ION, Beta-lactamase, ... | | Authors: | Powers, R.A, Mitchell, J.M, June, C.M. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
4Z98
 
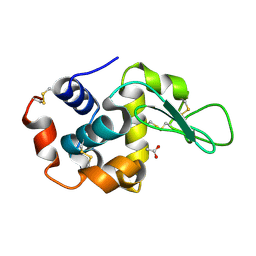 | | Crystal Structure of Hen Egg White Lysozyme using Serial X-ray Diffraction Data Collection | | Descriptor: | ACETATE ION, Lysozyme C | | Authors: | Murray, T.D, Lyubimov, A.Y, Ogata, C.M, Uervirojnangkoorn, M, Brunger, A.T, Berger, J.M. | | Deposit date: | 2015-04-10 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A high-transparency, micro-patternable chip for X-ray diffraction analysis of microcrystals under native growth conditions.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
7RPA
 
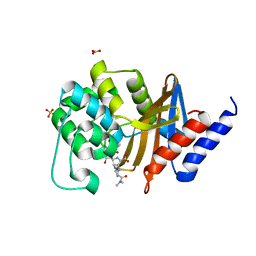 | | X-ray crystal structure of OXA-24/40 K84D in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, BICARBONATE ION, Beta-lactamase, ... | | Authors: | Powers, R.A, Mitchell, J.M, June, C.M. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
8F8L
 
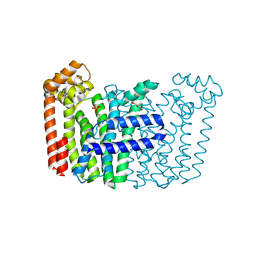 | | The structure of Rv2173 from M. tuberculosis with DMAP bound | | Descriptor: | (2E,6E)-farnesyl diphosphate synthase, CALCIUM ION, DIMETHYLALLYL DIPHOSPHATE, ... | | Authors: | Johnston, J.M, Allison, T.M, Titterington, J. | | Deposit date: | 2022-11-22 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of Rv2173 from M. tuberculosis in APO-, IPP-, and DMAP-bound forms.
To be Published
|
|
7RP8
 
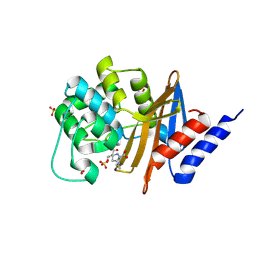 | |
4ZRN
 
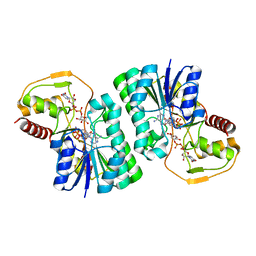 | |
7NHP
 
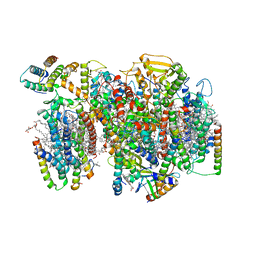 | | Structure of PSII-I (PSII with Psb27, Psb28, and Psb34) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Zabret, J, Bohn, S, Schuller, S.K, Arnolds, O, Chan, A, Tajkhorshid, E, Stoll, R, Engel, B.D, Rudack, T, Schuller, J.M, Nowaczyk, M.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural insights into photosystem II assembly.
Nat.Plants, 7, 2021
|
|
7NHO
 
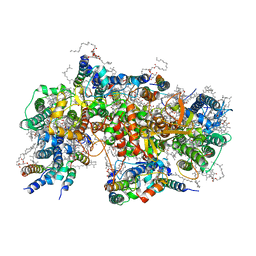 | | Structure of PSII-M | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Zabret, J, Bohn, S, Schuller, S.K, Arnolds, O, Chan, A, Tajkhorshid, E, Stoll, R, Engel, B.D, Rudack, T, Schuller, J.M, Nowaczyk, M.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural insights into photosystem II assembly.
Nat.Plants, 7, 2021
|
|
4ZI9
 
 | | Structure of mouse clustered PcdhgA1 EC1-3 | | Descriptor: | CALCIUM ION, MCG133388, isoform CRA_t | | Authors: | Nicoludis, J.M, Lau, S.-Y, Scharfe, C.P.I, Marks, D.S, Weihofen, W.A, Gaudet, R. | | Deposit date: | 2015-04-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Sequence Analyses of Clustered Protocadherins Reveal Antiparallel Interactions that Mediate Homophilic Specificity.
Structure, 23, 2015
|
|
6VEB
 
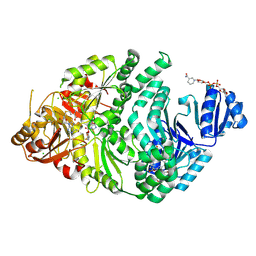 | | Precorrin-2-bound S128A S. typhimurium siroheme synthase | | Descriptor: | 3,3',3'',3'''-[(7S,8S,12S,13S)-3,8,13,17-tetrakis(carboxymethyl)-8,13-dimethyl-7,8,12,13,20,24-hexahydroporphyrin-2,7,1 2,18-tetrayl]tetrapropanoic acid, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Pennington, J.M, Stroupe, M.E. | | Deposit date: | 2019-12-31 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site.
Nat Commun, 11, 2020
|
|
5W0I
 
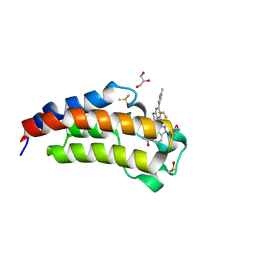 | | CREBBP Bromodomain in complex with Cpd8 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydrofuran-3-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-[(3S)-oxolan-3-yl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
7N06
 
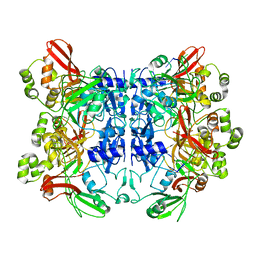 | | SARS-CoV-2 Nsp15 endoribonuclease post-cleavage state | | Descriptor: | RNA (5'-R(*AP*UP*A)-3'), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Dillard, L.B, Krahn, J.M, Stanley, R.E. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U.
Nucleic Acids Res., 49, 2021
|
|
7R6R
 
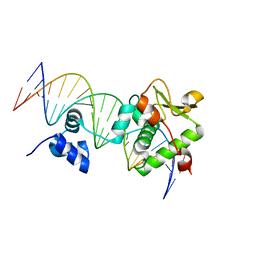 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
7NHQ
 
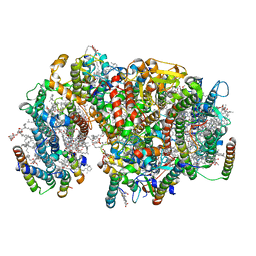 | | Structure of PSII-I prime (PSII with Psb28, and Psb34) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Zabret, J, Bohn, S, Schuller, S.K, Arnolds, O, Chan, A, Tajkhorshid, E, Stoll, R, Engel, B.D, Rudack, T, Schuller, J.M, Nowaczyk, M.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural insights into photosystem II assembly.
Nat.Plants, 7, 2021
|
|
4Z87
 
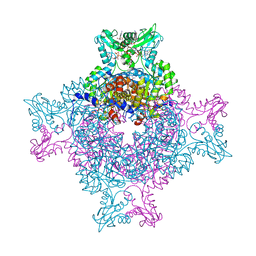 | | Structure of the IMP dehydrogenase from Ashbya gossypii bound to GDP | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Buey, R.M, de Pereda, J.M, Revuelta, J.L. | | Deposit date: | 2015-04-08 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases.
Nat Commun, 6, 2015
|
|
7MGM
 
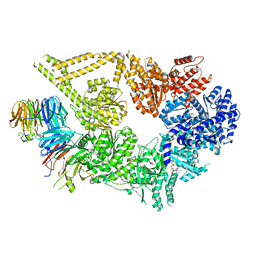 | | Structure of yeast cytoplasmic dynein with AAA3 Walker B mutation bound to Lis1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lahiri, I, Reimer, J.M, Leschziner, A.E. | | Deposit date: | 2021-04-12 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for cytoplasmic dynein-1 regulation by Lis1.
Elife, 11, 2022
|
|
6VJK
 
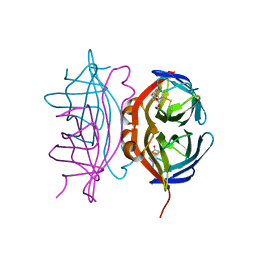 | | Streptavidin mutant M88 (N49C/A86C) | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Marangoni, J.M, Wu, S.C, Fogen, D, Wong, S.L, Ng, K.K.S. | | Deposit date: | 2020-01-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering a disulfide-gated switch in streptavidin enables reversible binding without sacrificing binding affinity.
Sci Rep, 10, 2020
|
|
4ZIR
 
 | | Crystal structure of EcfAA' heterodimer bound to AMPPNP | | Descriptor: | CHLORIDE ION, Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, ... | | Authors: | Karpowich, N.K, Cocco, N, Song, J.M, Wang, D.N. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ATP binding drives substrate capture in an ECF transporter by a release-and-catch mechanism.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6VKQ
 
 | | Crystal Structure of human PARP-1 CAT domain bound to inhibitor EB-47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Steffen, J.D, Pascal, J.M. | | Deposit date: | 2020-01-21 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for allosteric PARP-1 retention on DNA breaks.
Science, 368, 2020
|
|
7XLD
 
 | | Crystal structure of IsdH linker-NEAT3 bound to a nanobody (VHH) | | Descriptor: | Iron-regulated surface determinant protein H, MAGNESIUM ION, Nanobody VHH6, ... | | Authors: | Caaveiro, J.M.M, Valenciano-Bellido, S, Tsumoto, K. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting hemoglobin receptors IsdH and IsdB of Staphylococcus aureus with a single VHH antibody inhibits bacterial growth.
J.Biol.Chem., 299, 2023
|
|
4ZI8
 
 | | Structure of mouse clustered PcdhgC3 EC1-3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Nicoludis, J.M, Lau, S.-Y, Scharfe, C.P.I, Marks, D.S, Weihofen, W.A, Gaudet, R. | | Deposit date: | 2015-04-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structure and Sequence Analyses of Clustered Protocadherins Reveal Antiparallel Interactions that Mediate Homophilic Specificity.
Structure, 23, 2015
|
|
6VKK
 
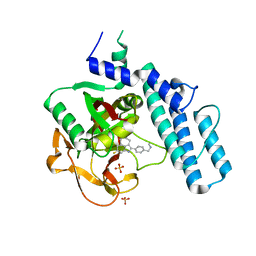 | |
7XT0
 
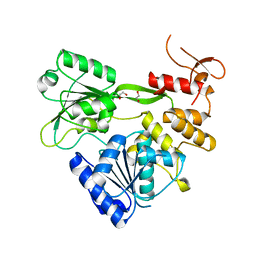 | | Crystal structure of RNA helicase from Saint Louis encephalitis virus and discovery of its inhibitors | | Descriptor: | 1,2-ETHANEDIOL, RNA helicase | | Authors: | Wang, D.P, Jiang, F.Y, Zeng, X.Y, Zhao, R, Chen, C, Zhu, Y, Cao, J.M. | | Deposit date: | 2022-05-15 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of RNA helicase from Saint Louis encephalitis virus and discovery of its inhibitors
To Be Published
|
|
7RFG
 
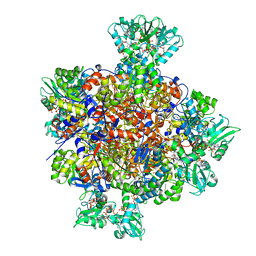 | | HUMAN IMPDH1 TREATED WITH GTP, IMP, AND NAD+ OCTAMER-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RFE
 
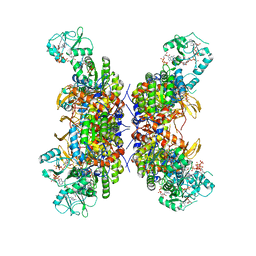 | | HUMAN IMPDH1 TREATED WITH GTP, IMP, AND NAD+; INTERFACE-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
