1OTC
 
 | | THE O. NOVA TELOMERE END BINDING PROTEIN COMPLEXED WITH SINGLE STRAND DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3'), PROTEIN (TELOMERE-BINDING PROTEIN ALPHA SUBUNIT), PROTEIN (TELOMERE-BINDING PROTEIN BETA SUBUNIT) | | Authors: | Horvath, M.P, Schweiker, V.L, Bevilacqua, J.M, Ruggles, J.A, Schultz, S.C. | | Deposit date: | 1998-11-25 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA.
Cell(Cambridge,Mass.), 95, 1998
|
|
3I15
 
 | |
3CNB
 
 | | Crystal structure of signal receiver domain of DNA binding response regulator protein (merR) from Colwellia psychrerythraea 34H | | Descriptor: | DNA-binding response regulator, merR family | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Hu, S, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of signal receiver domain of DNA binding response regulator (merR) from Colwellia psychrerythraea 34H.
To be Published
|
|
1P54
 
 | |
3Q6T
 
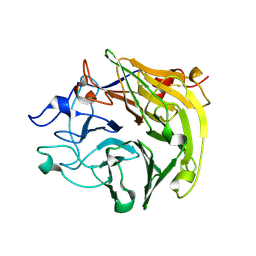 | | Salivary protein from Lutzomyia longipalpis, Ligand free | | Descriptor: | 43.2 kDa salivary protein, CITRIC ACID | | Authors: | Andersen, J.F, Xu, X, Chang, B.W, Collin, N, Valenzuela, J.G, Ribeiro, J.M. | | Deposit date: | 2011-01-03 | | Release date: | 2011-07-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structure and function of a "yellow" protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection.
J.Biol.Chem., 286, 2011
|
|
1P8K
 
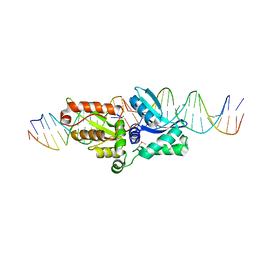 | |
3RHN
 
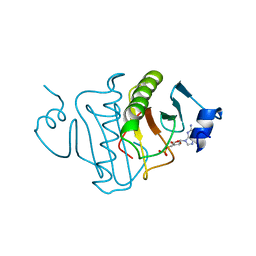 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT COMPLEXED WITH GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
3R09
 
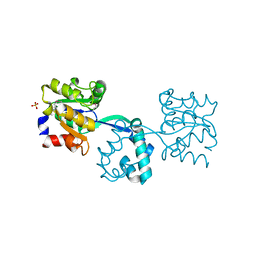 | | Crystal structure of probable HAD family hydrolase from Pseudomonas fluorescens Pf-5 with bound Mg | | Descriptor: | Hydrolase, haloacid dehalogenase-like family, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-03-07 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of probable HAD family hydrolase from Pseudomonas fluorescens Pf-5 with bound Mg
To be Published
|
|
1S7Z
 
 | | Structure of Ocr from Bacteriophage T7 | | Descriptor: | CESIUM ION, Gene 0.3 protein | | Authors: | Walkinshaw, M.D, Taylor, P, Sturrock, S.S, Atanasiu, C, Berg, T, Henderson, R.M, Edwardson, J.M, Dryden, D.T. | | Deposit date: | 2004-01-30 | | Release date: | 2004-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of Ocr from Bacteriophage T7, a Protein that Mimics B-Form DNA
Mol.Cell, 9, 2002
|
|
1N83
 
 | | Crystal Structure of the complex between the Orphan Nuclear Hormone Receptor ROR(alpha)-LBD and Cholesterol | | Descriptor: | CHOLESTEROL, Nuclear receptor ROR-alpha | | Authors: | Kallen, J.A, Schlaeppi, J.M, Bitsch, F, Geisse, S, Geiser, M, Delhon, I, Fournier, B. | | Deposit date: | 2002-11-19 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray Structure of hROR(alpha) LBD at 1.63A: Structural and Functional data that Cholesterol or a Cholesterol derivative is the natural ligand of ROR(alpha)
Structure, 10, 2002
|
|
1N8M
 
 | | Solution structure of Pi4, a four disulfide bridged scorpion toxin active on potassium channels | | Descriptor: | Potassium channel blocking toxin 4 | | Authors: | Guijarro, J.I, M'Barek, S, Olamendi-Portugal, T, Gomez-Lagunas, F, Garnier, D, Rochat, H, Possani, L.D, Sabatier, J.M, Delepierre, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-09-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pi4, a short four-disulfide-bridged scorpion toxin specific of potassium channels.
Protein Sci., 12, 2003
|
|
3I13
 
 | |
4BMK
 
 | | Serine Palmitoyltransferase K265A from S. paucimobilis with bound PLP- Myriocin Aldimine | | Descriptor: | Decarboxylated Myriocin, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wadsworth, J.M, Clarke, D.J, McMahon, S.A, Beattie, A.E, Lowther, J, Dunn, T.M, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Chemical Basis of Serine Palmitoyltransferase Inhibition by Myriocin.
J.Am.Chem.Soc., 135, 2013
|
|
3I45
 
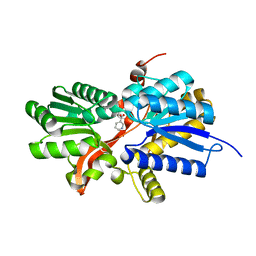 | | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum Atcc 11170 | | Descriptor: | NICOTINIC ACID, Twin-arginine translocation pathway signal protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum
Atcc 11170
To be Published
|
|
1N9E
 
 | | Crystal structure of Pichia pastoris Lysyl Oxidase PPLO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Guss, J.M, Duff, A.P. | | Deposit date: | 2002-11-24 | | Release date: | 2004-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Pichia pastoris Lysyl Oxidase
Biochemistry, 42, 2003
|
|
3R4G
 
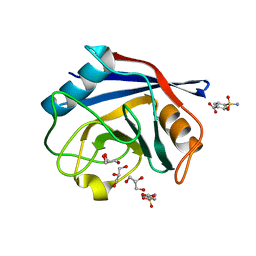 | | Human Cyclophilin D Complexed with a Fragment | | Descriptor: | 4-sulfamoylbenzoic acid, GLYCEROL, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | Deposit date: | 2011-03-17 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
3R57
 
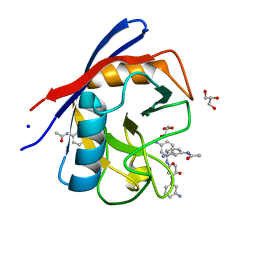 | | Human Cyclophilin D Complexed with a Fragment | | Descriptor: | GLYCEROL, N-(3-aminophenyl)acetamide, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | Deposit date: | 2011-03-18 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
3EUB
 
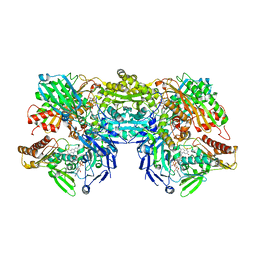 | | Crystal Structure of Desulfo-Xanthine Oxidase with Xanthine | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, HYDROXY(DIOXO)MOLYBDENUM, ... | | Authors: | Pauff, J.M, Cao, H, Hille, R. | | Deposit date: | 2008-10-09 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate Orientation and Catalysis at the Molybdenum Site in Xanthine Oxidase: CRYSTAL STRUCTURES IN COMPLEX WITH XANTHINE AND LUMAZINE.
J.Biol.Chem., 284, 2009
|
|
3ES7
 
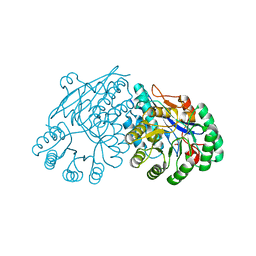 | | Crystal structure of divergent enolase from Oceanobacillus Iheyensis complexed with Mg and L-malate. | | Descriptor: | (2S)-2-hydroxybutanedioic acid, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-04 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
3EVM
 
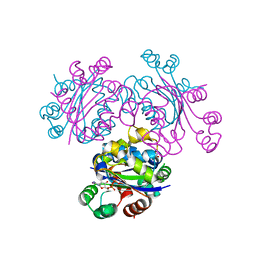 | | Crystal structure of the Mimivirus NDK +Kpn-N62L-R107G triple mutant complexed with dCDP | | Descriptor: | DEOXYCYTIDINE DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-10-13 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
1SNU
 
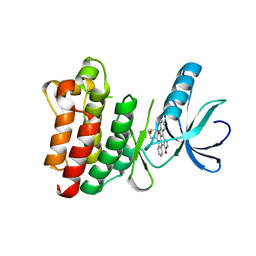 | | CRYSTAL STRUCTURE OF THE UNPHOSPHORYLATED INTERLEUKIN-2 TYROSINE KINASE CATALYTIC DOMAIN | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
3ETR
 
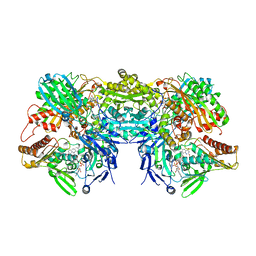 | | Crystal structure of xanthine oxidase in complex with lumazine | | Descriptor: | CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Pauff, J.M, Cao, H, Hille, R. | | Deposit date: | 2008-10-08 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate Orientation and Catalysis at the Molybdenum Site in Xanthine Oxidase: CRYSTAL STRUCTURES IN COMPLEX WITH XANTHINE AND LUMAZINE.
J.Biol.Chem., 284, 2009
|
|
4BWY
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI8 (R32) | | Descriptor: | P4 | | Authors: | El Omari, K, Meier, C, Kainov, D, Sutton, G, Grimes, J.M, Poranen, M.M, Bamford, D.H, Tuma, R, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-07-05 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Tracking in Atomic Detail the Functional Specializations in Viral Reca Helicases that Occur During Evolution.
Nucleic Acids Res., 41, 2013
|
|
1SFE
 
 | | ADA O6-METHYLGUANINE-DNA METHYLTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | ADA O6-METHYLGUANINE-DNA METHYLTRANSFERASE | | Authors: | Moore, M.H, Gulbis, J.M, Dodson, E.J, Demple, B, Moody, P.C.E. | | Deposit date: | 1996-06-21 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a suicidal DNA repair protein: the Ada O6-methylguanine-DNA methyltransferase from E. coli.
EMBO J., 13, 1994
|
|
4C04
 
 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with inhibitor | | Descriptor: | PROTEIN ARGININE N-METHYLTRANSFERASE 6, SINEFUNGIN | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
