6WU3
 
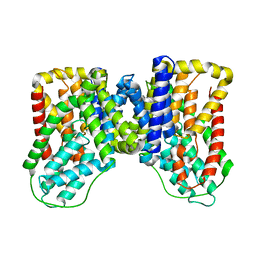 | |
4EDT
 
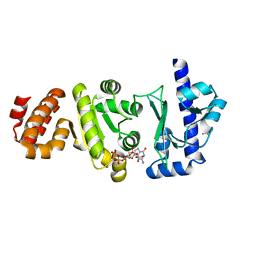 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to ppGpp and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4EDR
 
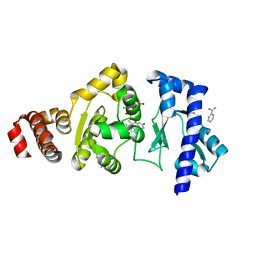 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to UTP and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, MANGANESE (II) ION, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
1G6Q
 
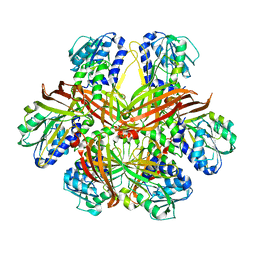 | | CRYSTAL STRUCTURE OF YEAST ARGININE METHYLTRANSFERASE, HMT1 | | Descriptor: | HNRNP ARGININE N-METHYLTRANSFERASE | | Authors: | Weiss, V.H, McBride, A.E, Soriano, M.A, Filman, D.J, Silver, P.A, Hogle, J.M. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure and oligomerization of the yeast arginine methyltransferase, Hmt1.
Nat.Struct.Biol., 7, 2000
|
|
4HUP
 
 | | Structure of ricin A chain bound with N-(N-(pterin-7-yl)carbonylglycyl)-L-phenylalanyl)-L-phenylalanine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-1H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-3-phenyl-propanoic acid, MALONIC ACID, Ricin, ... | | Authors: | Jasheway, K.R, Monzingo, A.F, Saito, R, Pruet, J.M, Manzano, L.A, Wiget, P.A, Anslyn, E.V, Robertus, J.D. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Peptide-conjugated pterins as inhibitors of ricin toxin A.
J.Med.Chem., 56, 2013
|
|
3TY4
 
 | |
2PNE
 
 | | Crystal Structure of the Snow Flea Antifreeze Protein | | Descriptor: | 6.5 kDa glycine-rich antifreeze protein | | Authors: | Pentelute, B.L, Kent, S.B.H, Gates, Z.P, Tereshko, V, Kossiakoff, A.A, Kurutz, J, Dashnau, J, Vaderkooi, J.M. | | Deposit date: | 2007-04-24 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers
J.Am.Chem.Soc., 130, 2008
|
|
4EDV
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to pppGpp and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, MANGANESE (II) ION, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
3TO1
 
 | | Two surfaces on Rtt106 mediate histone binding and chaperone activity | | Descriptor: | Histone chaperone RTT106 | | Authors: | Zunder, R.M, Antczak, A.J, Berger, J.M, Rine, J. | | Deposit date: | 2011-09-02 | | Release date: | 2011-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UA7
 
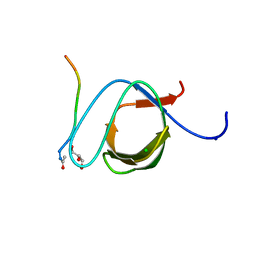 | | Crystal Structure of the Human Fyn SH3 domain in complex with a peptide from the Hepatitis C virus NS5A-protein | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Martin-Garcia, J.M, Ruiz-Sanz, J, Luque, I, Camara-Artigas, A. | | Deposit date: | 2011-10-21 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The promiscuous binding of the Fyn SH3 domain to a peptide from the NS5A protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4EER
 
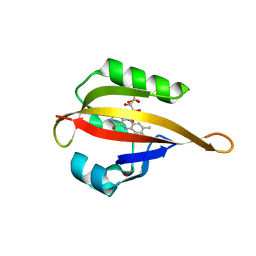 | | Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 C426A mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4ERS
 
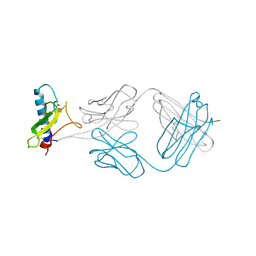 | | A Molecular Basis for Negative Regulation of the Glucagon Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Murray, J.M, Koth, C.M, Mukund, S. | | Deposit date: | 2012-04-20 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.637 Å) | | Cite: | Molecular basis for negative regulation of the glucagon receptor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6OQI
 
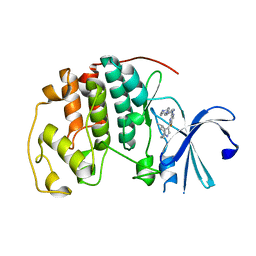 | | CDK2 in complex with Cpd14 (5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)-N-(5-(4-methylpiperazin-1-yl)pyridin-2-yl)pyrimidin-2-amine) | | Descriptor: | 5-fluoro-N-[5-(4-methylpiperazin-1-yl)pyridin-2-yl]-4-[(4S)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Murray, J.M. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3L25
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3UAN
 
 | | Crystal structure of 3-O-sulfotransferase (3-OST-1) with bound PAP and heptasaccharide substrate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Moon, A.F, Xu, Y, Woody, S.M, Krahn, J.M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ESI
 
 | | Structure of ricin A chain bound with N-((1H-1,2,3-triazol-4-yl)methyl-2-amino-4-oxo-3,4-dihydropteridine-7-carboxamide | | Descriptor: | 2-amino-4-oxo-N-(1H-1,2,3-triazol-5-ylmethyl)-1,4-dihydropteridine-7-carboxamide, Ricin | | Authors: | Jasheway, K.R, Pruet, J.M, Ryoto, S, Manzano, L.A, Wiget, P.A, Kamat, I, Anslyn, E.V, Monzingo, A.F, Robertus, J.D. | | Deposit date: | 2012-04-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Optimized 5-membered heterocycle-linked pterins for the inhibition of Ricin Toxin A.
ACS Med Chem Lett, 3, 2012
|
|
3TNZ
 
 | | Crystal structure of Mus musculus iodotyrosine deiodinase (IYD) C217A, C239A bound to FMN and mono-iodotyrosine (MIT) | | Descriptor: | 3-IODO-TYROSINE, CITRATE ANION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Buss, J.M, McTamney, P.M, Rokita, S.E. | | Deposit date: | 2011-09-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Expression of a soluble form of iodotyrosine deiodinase for active site characterization by engineering the native membrane protein from Mus musculus.
Protein Sci., 21, 2012
|
|
4HV7
 
 | | Structure of ricin A chain bound with N-(N-(pterin-7-yl)carbonylglycyl)glycine | | Descriptor: | 2-[2-[(2-azanyl-4-oxidanylidene-1H-pteridin-7-yl)carbonylamino]ethanoylamino]ethanoic acid, MALONIC ACID, Ricin, ... | | Authors: | Jasheway, K.R, Monzingo, A.F, Saito, R, Pruet, J.M, Manzano, L.A, Wiget, P.A, Anslyn, E.V, Robertus, J.D. | | Deposit date: | 2012-11-05 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.869 Å) | | Cite: | Peptide-conjugated pterins as inhibitors of ricin toxin A.
J.Med.Chem., 56, 2013
|
|
4HJK
 
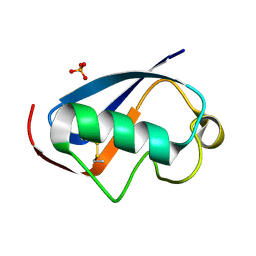 | | U7Ub7 Disulfide variant | | Descriptor: | PHOSPHATE ION, Ubiquitin | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2012-10-12 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Potent and selective inhibitors of USP7/HAUSP activity by protein conformational stabilization
to be published
|
|
4EDK
 
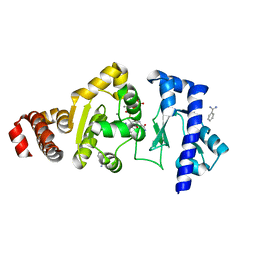 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to GTP and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4EEP
 
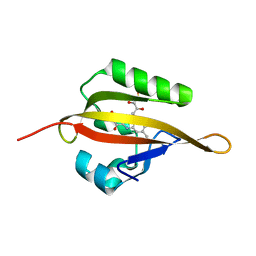 | | Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EKJ
 
 | | Crystal structure of a monomeric beta-xylosidase from Caulobacter crescentus CB15 | | Descriptor: | Beta-xylosidase, SULFATE ION | | Authors: | Santos, C.R, Polo, C.C, Correa, J.M, Simao, R.C.G, Seixas, F.A.V, Murakami, M.T. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The accessory domain changes the accessibility and molecular topography of the catalytic interface in monomeric GH39 beta-xylosidases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4I1K
 
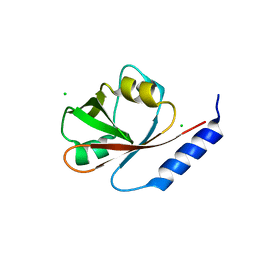 | | Crystal Structure of VRN1 (Residues 208-341) | | Descriptor: | B3 domain-containing transcription factor VRN1, CHLORIDE ION | | Authors: | King, G, Chanson, A.H, McCallum, E.J, Ohme-Takagi, M, Byriel, K, Hill, J.M, Martin, J.L, Mylne, J.S. | | Deposit date: | 2012-11-21 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Arabidopsis B3 Domain Protein VERNALIZATION1 (VRN1) Is Involved in Processes Essential for Development, with Structural and Mutational Studies Revealing Its DNA-binding Surface.
J.Biol.Chem., 288, 2013
|
|
3TO0
 
 | | Crystal structure of Mus musculus iodotyrosine deiodinase (IYD) C217A, C239A bound to FMN | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Iodotyrosine deiodinase 1, ... | | Authors: | Buss, J.M, McTamney, P.M, Rokita, S.E. | | Deposit date: | 2011-09-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Expression of a soluble form of iodotyrosine deiodinase for active site characterization by engineering the native membrane protein from Mus musculus.
Protein Sci., 21, 2012
|
|
4ESJ
 
 | | RESTRICTION ENDONUCLEASE DpnI IN COMPLEX WITH TARGET DNA | | Descriptor: | AZIDE ION, DNA (5'-D(*CP*TP*GP*GP*(6MA)P*TP*CP*CP*AP*G)-3'), GLYCEROL, ... | | Authors: | Siwek, W, Czapinska, H, Bochtler, M, Bujnicki, J.M, Skowronek, K. | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure and mechanism of action of the N6-methyladenine-dependent type IIM restriction endonuclease R.DpnI.
Nucleic Acids Res., 40, 2012
|
|
