5YD4
 
 | |
8QZD
 
 | | Soluble epoxide hydrolase in complex with Epoxykinin | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-bromanyl-3-[2,2,2-tris(fluoranyl)ethanoyl]indol-1-yl]-N-cycloheptyl-ethanamide, BROMIDE ION, ... | | Authors: | Kumar, A, Ehrler, J.M.H, Ziegler, S, Doetsch, L, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of the sEH Inhibitor Epoxykynin as a Potent Kynurenine Pathway Modulator.
J.Med.Chem., 67, 2024
|
|
1TZY
 
 | | Crystal Structure of the Core-Histone Octamer to 1.90 Angstrom Resolution | | Descriptor: | CHLORIDE ION, HISTONE H3, HISTONE H4-VI, ... | | Authors: | Wood, C.M, Nicholson, J.M, Chantalat, L, Reynolds, C.D, Lambert, S.J, Baldwin, J.P. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of the native histone octamer.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1U86
 
 | |
5QDC
 
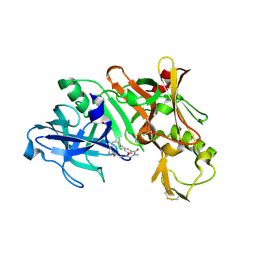 | | Crystal structure of BACE complex with BMC019 hydrolyzed | | Descriptor: | (4S)-4-[(1R)-1,2-dihydroxyethyl]-N,N-dimethyl-2-oxo-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaene-19-carboxamide, Beta-secretase 1, GLYCEROL | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1UCL
 
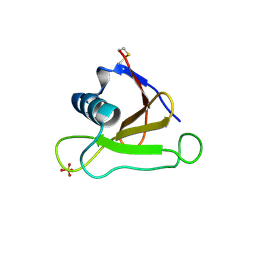 | | Mutants of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Takano, K, Scholtz, J.M, Sacchettini, J.C, Pace, C.N. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The contribution of polar group burial to protein stability is strongly context-dependent
J.Biol.Chem., 278, 2003
|
|
5QD2
 
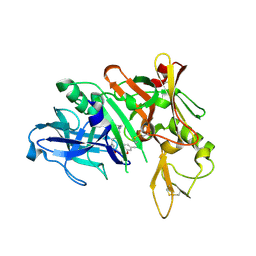 | | Crystal structure of BACE complex with BMC017 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-19-(methoxymethyl)-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5Y25
 
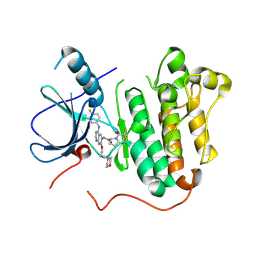 | | EGFR kinase domain mutant (T790M/L858R) with covalent ligand NS-062 | | Descriptor: | (2R)-N-[4-[(3-chloranyl-4-fluoranyl-phenyl)amino]-7-(3-morpholin-4-ylpropoxy)quinazolin-6-yl]-1-(2-fluoranylethanoyl)pyrrolidine-2-carboxamide, Epidermal growth factor receptor | | Authors: | Shiroishi, M, Abe, Y, Caaveiro, J.M.M, Sakamoto, S, Morimoto, S, Fuchida, H, Shindo, N, Ojida, A. | | Deposit date: | 2017-07-24 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Selective and reversible modification of kinase cysteines with chlorofluoroacetamides.
Nat.Chem.Biol., 15, 2019
|
|
1TXY
 
 | | E. coli PriB | | Descriptor: | Primosomal replication protein n | | Authors: | Keck, J.L, Lopper, M, Holton, J.M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PriB, a component of the Escherichia coli replication restart primosome
Structure, 12, 2004
|
|
5GHW
 
 | | Crystal structure of broad neutralizing antibody 10E8 with long epitope bound | | Descriptor: | Endogenous retrovirus group K member 8 Env polyprotein, FAB 10E8 HEAVY CHAIN, FAB 10E8 LIGHT CHAIN, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Morante, K, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2016-06-21 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface
Sci Rep, 6, 2016
|
|
5QDA
 
 | | Crystal structure of BACE complex with BMC013 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-18-methoxy-3,15,17-triazatricyclo[14.3.1.1~6,10~]henicosa-1(20),6(21),7,9,16,18-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCO
 
 | | Crystal structure of BACE complex with BMC016 | | Descriptor: | (4S)-19-acetyl-4-[(1R)-1-hydroxy-2-({1-[3-(propan-2-yl)phenyl]cyclopropyl}amino)ethyl]-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1TOE
 
 | | Unliganded structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, SULFATE ION | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
5QD3
 
 | | Crystal structure of BACE complex with BMC010 | | Descriptor: | (10R,12S)-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-17-(methoxymethyl)-10-methyl-2,13-diazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1TUE
 
 | |
5QDD
 
 | | Crystal structure of BACE complex with BMC020 hydrolyzed | | Descriptor: | (10R,12S)-12-[(1R)-1,2-dihydroxyethyl]-N,N,10-trimethyl-14-oxo-2-oxa-13-azabicyclo[13.3.1]nonadeca-1(19),15,17-triene-17-carboxamide, Beta-secretase 1, GLYCEROL | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1TOG
 
 | | Hydrocinnamic acid-bound structure of SRHEPT + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
5QCY
 
 | | Crystal structure of BACE complex with BMC008 | | Descriptor: | (9R,11S)-11-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-9-methyl-16-(1,3-oxazol-2-yl)-3-[(1R)-1-phenylethyl]-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-triene-2,13-dione, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCX
 
 | | Crystal structure of BACE complex with BMC007 | | Descriptor: | (9R,11S)-3-ethyl-11-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-9-methyl-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-triene-2,13-dione, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD9
 
 | | Crystal structure of BACE complex with BMC005 | | Descriptor: | (5S,8S,10R)-8-[(1R)-2-{[1-(3-tert-butylphenyl)cyclopropyl]amino}-1-hydroxyethyl]-4,5,10-trimethyl-1-oxa-4,7-diazacyclohexadecane-3,6-dione, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCP
 
 | | Crystal structure of BACE complex with BMC018 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-19-(2-oxopropoxy)-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD5
 
 | | Crystal structure of BACE complex with BMC009 | | Descriptor: | (10S,12S)-17-chloro-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-10-methyl-7-oxa-2,13,18-triazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1TZG
 
 | | Crystal structure of HIV-1 neutralizing human Fab 4E10 in complex with a 13-residue peptide containing the 4E10 epitope on gp41 | | Descriptor: | Envelope polyprotein GP160, Fab 4E10, GLYCEROL | | Authors: | Cardoso, R.M.F, Zwick, M.B, Stanfield, R.L, Kunert, R, Binley, J.M, Katinger, H, Burton, D.R, Wilson, I.A. | | Deposit date: | 2004-07-09 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Broadly Neutralizing Anti-HIV Antibody 4E10 Recognizes a Helical Conformation of a Highly Conserved Fusion-Associated Motif in gp41
Immunity, 22, 2005
|
|
5YD5
 
 | |
5QCU
 
 | | Crystal structure of BACE complex with BMC022 | | Descriptor: | (2R,4S)-N-butyl-4-[(5S,8S,10R)-5,10-dimethyl-3,3,6-trioxo-3lambda~6~-thia-7-azabicyclo[11.3.1]heptadeca-1(17),13,15-trien-8-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
