4RHN
 
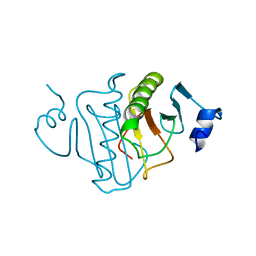 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT COMPLEXED WITH ADENOSINE | | Descriptor: | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN, alpha-D-ribofuranose | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-26 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
4RHZ
 
 | | Crystal structure of Cry23Aa1 and Cry37Aa1 binary protein complex | | Descriptor: | CALCIUM ION, Cry23AA1, Cry37AA1, ... | | Authors: | Rydel, T.J, Williams, J.M, Brown, G.R, Guzov, V.M, Sturman, E.J, Evdokimov, A. | | Deposit date: | 2014-10-03 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Associated Bacillus thuringiensis Binary Protein Complex of Cry23Aa1 and Cry37Aa1: Crystal Structure, Insecticidal Data, and Pore Formation Modeling.
To be Published
|
|
4RKP
 
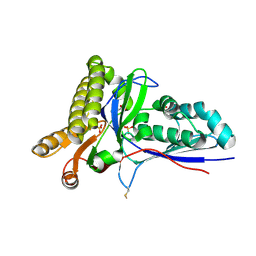 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (apo form) | | Descriptor: | ACETATE ION, Putative uncharacterized protein Ta1305, SULFATE ION | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
4RKZ
 
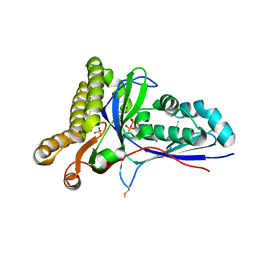 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (Mevalonate 3-Phosphate/ADP Bound) | | Descriptor: | (3R)-5-hydroxy-3-methyl-3-(phosphonooxy)pentanoic acid, ADENOSINE-5'-DIPHOSPHATE, Putative uncharacterized protein Ta1305, ... | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
4RSO
 
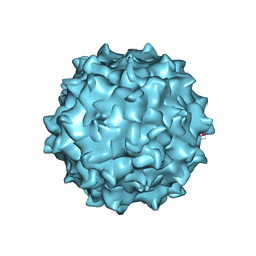 | | The structure of the neurotropic AAVrh.8 viral vector | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, CHLORIDE ION, Capsid protein VP1, ... | | Authors: | Halder, S, Van Vliet, K, Smith, J.K, McKenna, R, Wilson, J.M, Agbandje-McKenna, M. | | Deposit date: | 2014-11-10 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of neurotropic adeno-associated virus AAVrh.8.
J.Struct.Biol., 192, 2015
|
|
7OI3
 
 | | Cryo-EM structure of the Cetacean morbillivirus nucleoprotein-RNA complex | | Descriptor: | Cetacean morbillivirus nucleoprotein, poly-A 6-mer | | Authors: | Zinzula, L, Beck, F, Klumpe, S, Bohn, S, Pfeifer, G, Bollschweiler, D, Nagy, I, Plitzko, J.M, Baumeister, W. | | Deposit date: | 2021-05-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the cetacean morbillivirus nucleoprotein-RNA complex.
J.Struct.Biol., 213, 2021
|
|
7NT8
 
 | | Influenza virus H3N2 nucleoprotein - R416A mutant | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Nucleoprotein | | Authors: | Keown, J.R, Knight, M.L, Grimes, J.M, Fodor, E. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure of an H3N2 influenza virus nucleoprotein.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
4TN0
 
 | | Crystal Structure of the C-terminal Periplasmic Domain of Phosphoethanolamine Transferase EptC from Campylobacter jejuni | | Descriptor: | UPF0141 protein yjdB, ZINC ION | | Authors: | Fage, C.D, Brown, D, Boll, J.M, Keatinge-Clay, A.T, Trent, M.S. | | Deposit date: | 2014-06-02 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic study of the phosphoethanolamine transferase EptC required for polymyxin resistance and motility in Campylobacter jejuni.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4RM8
 
 | | Crystal structure of human ezrin in space group P21 | | Descriptor: | Ezrin | | Authors: | Phang, J.M, Harrop, S.J, Davies, R, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2014-10-20 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin.
Biochem. J., 473, 2016
|
|
4RMA
 
 | | Crystal structure of the FERM domain of human ezrin | | Descriptor: | Ezrin, SULFATE ION | | Authors: | Phang, J.M, Harrop, S.J, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2014-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin.
Biochem. J., 473, 2016
|
|
4S1K
 
 | | Structure of Uranotaenia sapphirina cypovirus (CPV17) polyhedrin at 100 K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Ginn, H.M, Messerschmidt, M, Ji, X, Zhang, H, Axford, D, Gildea, R.J, Winter, G, Brewster, A.S, Hattne, J, Wagner, A, Grimes, J.M, Evans, G, Sauter, N.K, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of CPV17 polyhedrin determined by the improved analysis of serial femtosecond crystallographic data.
Nat Commun, 6, 2015
|
|
7O82
 
 | | The L-arginine/agmatine antiporter from E. coli at 1.7 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine/agmatine antiporter, DECANE, ... | | Authors: | Jeckelmann, J.M, Ilgue, H, Kalbermatter, D, Fotiadis, D. | | Deposit date: | 2021-04-14 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution structure of the amino acid transporter AdiC reveals insights into the role of water molecules and networks in oligomerization and substrate binding.
Bmc Biol., 19, 2021
|
|
4RKS
 
 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (Mevalonate Bound) | | Descriptor: | (R)-MEVALONATE, ACETATE ION, GLYCEROL, ... | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
4RO5
 
 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM | | Descriptor: | GLYCEROL, SAT domain from CazM | | Authors: | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | Deposit date: | 2014-10-27 | | Release date: | 2015-09-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4ROS
 
 | |
4TMA
 
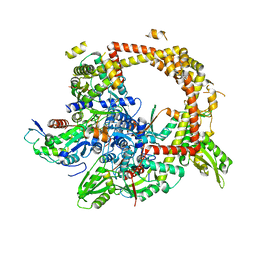 | | Crystal structure of gyrase bound to its inhibitor YacG | | Descriptor: | DNA gyrase inhibitor YacG, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Vos, S.M, Lyubimov, A.Y, Hershey, D.M, Schoeffler, A.J, Berger, J.M. | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Direct control of type IIA topoisomerase activity by a chromosomally encoded regulatory protein.
Genes Dev., 28, 2014
|
|
7O62
 
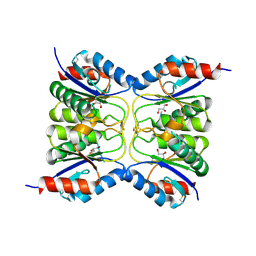 | |
4RM9
 
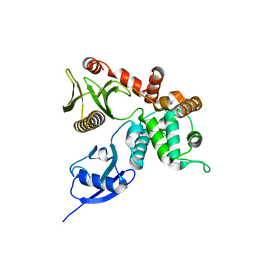 | | Crystal structure of human ezrin in space group C2221 | | Descriptor: | Ezrin | | Authors: | Phang, J.M, Harrop, S.J, Davies, R, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2014-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin.
Biochem. J., 473, 2016
|
|
4S13
 
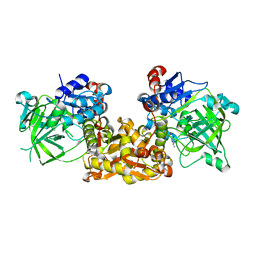 | | Ferulic Acid Decarboxylase (FDC1) | | Descriptor: | 4-ethenylphenol, Ferulic acid decarboxylase 1 | | Authors: | Lee, S.G, Bhuiya, M.W, Yu, O, Jez, J.M. | | Deposit date: | 2015-01-07 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Structure and Mechanism of Ferulic Acid Decarboxylase (FDC1) from Saccharomyces cerevisiae.
Appl.Environ.Microbiol., 81, 2015
|
|
7OJA
 
 | | SaFtsZ(D210N) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OHK
 
 | | SaFtsZ complexed with GDP, BeF3- and Mg2+ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-11 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OHH
 
 | | SaFtsZ complexed with GDP and BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-11 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OJC
 
 | | SaFtsZ(Q48A) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OHN
 
 | | SaFtsZ complexed with GDP, AlF4- and Mg2+ | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-11 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OJB
 
 | | SaFtsZ(R143K) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
