1T98
 
 | | Crystal Structure of MukF(1-287) | | Descriptor: | Chromosome partition protein mukF | | Authors: | Fennell-Fezzie, R, Berger, J.M. | | Deposit date: | 2004-05-14 | | Release date: | 2005-05-24 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The MukF subunit of Escherichia coli condensin: architecture and functional relationship to kleisins.
Embo J., 24, 2005
|
|
3ILW
 
 | | Structure of DNA gyrase subunit A N-terminal domain | | Descriptor: | DNA gyrase subunit A, GLYCEROL | | Authors: | Tretter, E.M, Schoeffler, A.J, Weisfield, S.R, Berger, J.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-08-07 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Crystal structure of the DNA gyrase GyrA N-terminal domain from Mycobacterium tuberculosis.
Proteins, 78, 2010
|
|
4CR2
 
 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3RKP
 
 | | Crystal structure of BcpA*(D312A), the major pilin subunit of Bacillus cereus | | Descriptor: | Collagen adhesion protein | | Authors: | Hendrickx, A.P, Poor, C.B, Jureller, J.E, Budzik, J.M, He, C, Schneewind, O. | | Deposit date: | 2011-04-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Isopeptide bonds of the major pilin protein BcpA influence pilus structure and bundle formation on the surface of Bacillus cereus.
Mol.Microbiol., 85, 2012
|
|
3R91
 
 | | Macrocyclic lactams as potent Hsp90 inhibitors with excellent tumor exposure and extended biomarker activity. | | Descriptor: | (6S)-4,6,15,15,18-pentamethyl-5,17-dioxo-2,3,4,5,6,7,14,15,16,17-decahydro-1H-12,8-(metheno)[1,4,9]triazacyclotetradecino[9,8-a]indole-9-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, McBean, J.L, Dushin, R.G, Nittoli, T, Otteng, M, Ingalls, C, Golas, J.M, Liu, H, Lucas, J, Boschelli, F, Vogan, E, Hu, Y, Levin, J.I. | | Deposit date: | 2011-03-24 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Macrocyclic lactams as potent Hsp90 inhibitors with excellent tumor exposure and extended biomarker activity.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
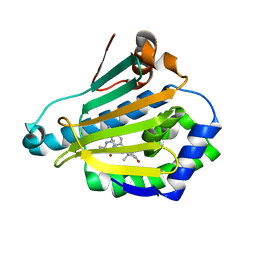
1P45
 
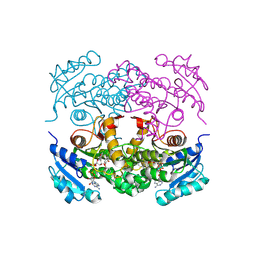 | | Targeting tuberculosis and malaria through inhibition of enoyl reductase: compound activity and structural data | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Kuo, M.R, Morbidoni, H.R, Alland, D, Sneddon, S.F, Gourlie, B.B, Staveski, M.M, Leonard, M, Gregory, J.S, Janjigian, A.D, Yee, C, Musser, J.M, Kreiswirth, B.N, Iwamoto, H, Perozzo, R, Jacobs Jr, W.R, Sacchettini, J.C, Fidock, D.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-21 | | Release date: | 2003-09-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeting tuberculosis and malaria through inhibition of Enoyl reductase: compound activity and structural data.
J.Biol.Chem., 278, 2003
|
|
4CR4
 
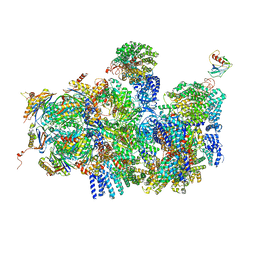 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2H51
 
 | |
3RTO
 
 | | Acoustically mounted porcine insulin microcrystals yield an X-ray SAD structure | | Descriptor: | Insulin, ZINC ION | | Authors: | Soares, A.S, Engel, M.A, Stearns, R, Datwani, S, Olechno, J, Ellson, R, Skinner, J.M, Allaire, M, Orville, A.M. | | Deposit date: | 2011-05-03 | | Release date: | 2011-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acoustically Mounted Microcrystals Yield High-Resolution X-ray Structures.
Biochemistry, 50, 2011
|
|
1R31
 
 | | HMG-CoA reductase from Pseudomonas mevalonii complexed with HMG-CoA | | Descriptor: | (R)-MEVALONATE, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, COENZYME A, ... | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-CoA Reductase.
To be Published
|
|
4CR3
 
 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ER6
 
 | | Crystal structure of a putative transcriptional regulator protein from Vibrio parahaemolyticus | | Descriptor: | Putative transcriptional regulator protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative transcriptional regulator protein from Vibrio parahaemolyticus
To be Published
|
|
3ERV
 
 | | Crystal structure of an putative C39-like peptidase from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, putative C39-like peptidase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an putative C39-like peptidase from Bacillus anthracis
To be Published
|
|
4D8L
 
 | | Crystal structure of the 2-pyrone-4,6-dicarboxylic acid hydrolase from sphingomonas paucimobilis | | Descriptor: | 2-pyrone-4,6-dicarbaxylate hydrolase | | Authors: | Malashkevich, V.N, Toro, R, Bonanno, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2012-01-10 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
3EDM
 
 | | Crystal structure of a short chain dehydrogenase from Agrobacterium tumefaciens | | Descriptor: | Short chain dehydrogenase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-03 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a short chain dehydrogenase from Agrobacterium tumefaciens
To be Published
|
|
1P7A
 
 | | Solution Structure of the Third Zinc Finger from BKLF | | Descriptor: | Kruppel-like factor 3, ZINC ION | | Authors: | Simpson, R.J.Y, Cram, E.D, Czolij, R, Matthews, J.M, Crossley, M, Mackay, J.P. | | Deposit date: | 2003-04-30 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | CCHX zinc finger derivatives retain the ability to bind Zn(II) and mediate protein-DNA interactions.
J.Biol.Chem., 278, 2003
|
|
2HE2
 
 | | Crystal structure of the 3rd PDZ domain of human discs large homologue 2, DLG2 | | Descriptor: | Discs large homolog 2 | | Authors: | Turnbull, A.P, Phillips, C, Berridge, G, Savitsky, P, Smee, C.E.A, Papagrigoriou, E, Debreczeni, J, Gorrec, F, Elkins, J.M, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
4D0Z
 
 | | GalNAc-T2 crystal soaked with UDP-5SGalNAc, mEA2 and manganese (Higher resolution dataset) | | Descriptor: | (2R,3R,4R,5R,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-5-thio-alpha-D-galactopyranose, ... | | Authors: | Lira-Navarrete, E, Iglesias-Fernandez, J, Zandberg, W.F, Companon, I, Kong, Y, Corzana, F, Pinto, B.M, Clausen, H, Peregrina, J.M, Vocadlo, D, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2014-04-30 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-Guided Front-Face Reaction Revealed by Combined Structural Snapshots and Metadynamics for the Polypeptide N- Acetylgalactosaminyltransferase 2.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3EGC
 
 | | Crystal structure of a putative ribose operon repressor from Burkholderia thailandensis | | Descriptor: | putative ribose operon repressor | | Authors: | Bonanno, J.B, Patskovsky, Y, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a putative ribose operon repressor from Burkholderia thailandensis
To be Published
|
|
4D9T
 
 | | Rsk2 C-terminal Kinase Domain with inhibitor (E)-methyl 3-(4-amino-7-(3-hydroxypropyl)-5-p-tolyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-2-cyanoacrylate | | Descriptor: | Ribosomal protein S6 kinase alpha-3, SODIUM ION, methyl (2S)-3-{4-amino-7-[(1E)-3-hydroxyprop-1-en-1-yl]-5-(4-methylphenyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl}-2-cyanopropanoate | | Authors: | Serafimova, I.M, Pufall, M.A, Krishnan, S, Duda, K, Cohen, M.S, Maglathlin, R.L, McFarland, J.M, Miller, R.M, Frodin, M, Taunton, J. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-25 | | Last modified: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles.
Nat.Chem.Biol., 8, 2012
|
|
3EMX
 
 | | Crystal structure of thioredoxin from Aeropyrum pernix | | Descriptor: | Thioredoxin | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of thioredoxin from Aeropyrum pernix
To be Published
|
|
1OMZ
 
 | | crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) in complex with UDPGalNAc | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2),
a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1RKY
 
 | | PPLO + Xe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Guss, J.M, Duff, A.P. | | Deposit date: | 2003-11-24 | | Release date: | 2004-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Using Xenon as a Probe for Dioxygen-binding Sites in Copper Amine Oxidases
J.Mol.Biol., 344, 2004
|
|
1OW6
 
 | | Paxillin LD4 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Campbell, I.D, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular Recognition of Paxillin LD motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
3ETM
 
 | | Crystal structure of the mimivirus NDK +KPN-N62L-R107G triple mutant complexed with CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-10-08 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
