2P67
 
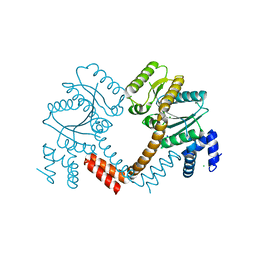 | | Crystal structure of LAO/AO transport system kinase | | Descriptor: | CHLORIDE ION, LAO/AO transport system kinase, SODIUM ION | | Authors: | Ramagopal, U.A, Adams, J, Rodgers, L, Toro, R, Bain, K, Rutter, M, Schwinn, K, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of LAO/AO transport system kinase
To be Published
|
|
2ZAY
 
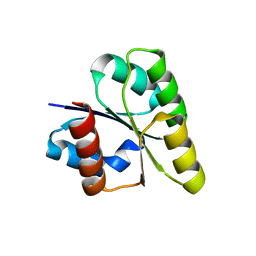 | | Crystal structure of response regulator from Desulfuromonas acetoxidans | | Descriptor: | Response regulator receiver protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of response regulator from Desulfuromonas acetoxidans.
To be Published
|
|
1IDC
 
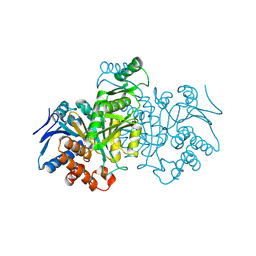 | | ISOCITRATE DEHYDROGENASE FROM E.COLI (MUTANT K230M), STEADY-STATE INTERMEDIATE COMPLEX DETERMINED BY LAUE CRYSTALLOGRAPHY | | Descriptor: | 2-OXALOSUCCINIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
3JU2
 
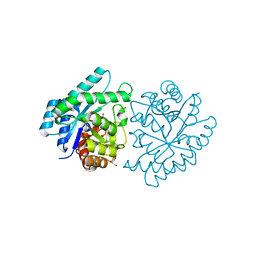 | | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, ZINC ION, uncharacterized protein SMc04130 | | Authors: | Patskovsky, Y, Foti, R, Ramagopal, U, Malashkevich, V, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti
To be Published
|
|
2C63
 
 | | 14-3-3 Protein Eta (Human) Complexed to Peptide | | Descriptor: | 14-3-3 PROTEIN ETA, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | Authors: | Elkins, J.M, Yang, X, Smee, C.E.A, Johansson, C, Sundstrom, M, Edwards, A, Weigelt, J, Arrowsmith, C, Doyle, D.A. | | Deposit date: | 2005-11-07 | | Release date: | 2005-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2VHC
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 N234G mutant in complex with AMPCPP and MN | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
1IDF
 
 | | ISOCITRATE DEHYDROGENASE K230M MUTANT APO ENZYME | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
2Z8O
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | 27.5 kDa virulence protein, L(+)-TARTARIC ACID | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1IE8
 
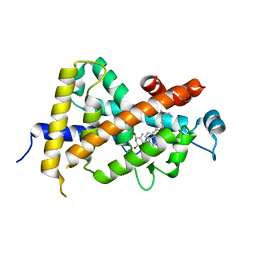 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060 | | Descriptor: | 5-(2-{1-[1-(4-ETHYL-4-HYDROXY-HEXYLOXY)-ETHYL]-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE}-ETHYLIDENE)-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2Z3S
 
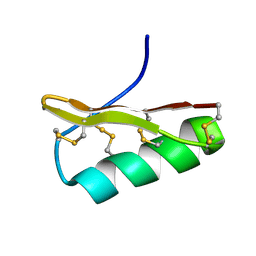 | | NMR structure of AgTx2-MTX | | Descriptor: | AgTx2-MTX | | Authors: | Pimentel, C, M'Barrek, S, Visan, V, Grissmer, S, Sabatier, J.M, Darbon, H, Fajloun, Z. | | Deposit date: | 2007-06-06 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Chemical synthesis and 1H-NMR 3D structure determination of AgTx2-MTX chimera, a new potential blocker for Kv1.2 channel, derived from MTX and AgTx2 scorpion toxins.
Protein Sci., 17, 2008
|
|
2BOX
 
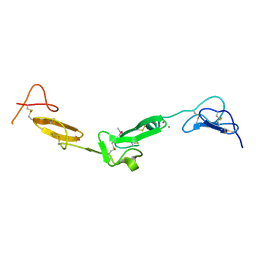 | | EGF Domains 1,2,5 of human EMR2, a 7-TM immune system molecule, in complex with strontium. | | Descriptor: | CACODYLATE ION, CHLORIDE ION, EGF-LIKE MODULE CONTAINING MUCIN-LIKE HORMONE RECEPTOR-LIKE 2 PRECURSOR, ... | | Authors: | Abbott, R.J.M, Spendlove, I, Roversi, P, Teriete, P, Knott, V, Handford, P.A, McDonnell, J.M, Lea, S.M. | | Deposit date: | 2005-04-14 | | Release date: | 2006-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of a Novel T Cell Receptor Co-Regulatory Protein Complex, Cd97-Cd55.
J.Biol.Chem., 282, 2007
|
|
2V3X
 
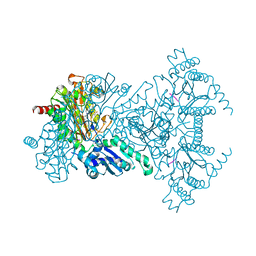 | |
1IH1
 
 | |
1IH6
 
 | |
2V1Y
 
 | | Structure of a phosphoinositide 3-kinase alpha adaptor-binding domain (ABD) in a complex with the iSH2 domain from p85 alpha | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT ALPHA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM | | Authors: | Miled, N, Yan, Y, Hon, W.C, Perisic, O, Zvelebil, M, Inbar, Y, Schneidman-Duhovny, D, Wolfson, H.J, Backer, J.M, Williams, R.L. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Two Classes of Cancer Mutations in the Phosphoinositide 3-Kinase Catalytic Subunit.
Science, 317, 2007
|
|
2BWX
 
 | | His354Ala Escherichia coli Aminopeptidase P | | Descriptor: | AMINOPEPTIDASE P, CHLORIDE ION, MANGANESE (II) ION | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BYG
 
 | | 2nd PDZ Domain of Discs Large Homologue 2 | | Descriptor: | CHANNEL ASSOCIATED PROTEIN OF SYNAPSE-110 | | Authors: | Elkins, J.M, Schoch, G.A, Smee, C.E.A, Berridge, G, Salah, E, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Pick1 and Other Pdz Domains Obtained with the Help of Self-Binding C-Terminal Extensions.
Protein Sci., 16, 2007
|
|
2BWU
 
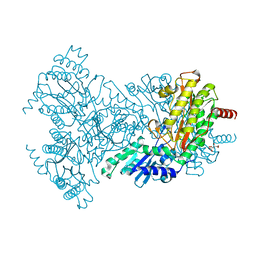 | | Asp271Ala Escherichia coli Aminopeptidase P | | Descriptor: | AMINOPEPTIDASE P, CITRATE ANION, MAGNESIUM ION, ... | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
4ZML
 
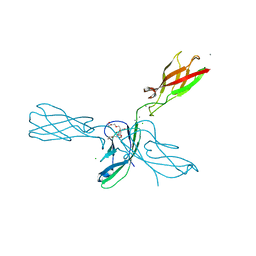 | | Crystal structure of human P-cadherin (ss-dimer) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cadherin-3, ... | | Authors: | Caaveiro, J.M.M, Kudo, S, Tsumoto, K. | | Deposit date: | 2015-05-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Adhesive Dimerization of Human P-Cadherin Catalyzed by a Chaperone-like Mechanism
Structure, 24, 2016
|
|
4ZMV
 
 | | Crystal structure of human P-cadherin (ss-X-dimer pocket I) | | Descriptor: | CALCIUM ION, Cadherin-3, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kudo, S, Tsumoto, K. | | Deposit date: | 2015-05-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Adhesive Dimerization of Human P-Cadherin Catalyzed by a Chaperone-like Mechanism
Structure, 24, 2016
|
|
3K4E
 
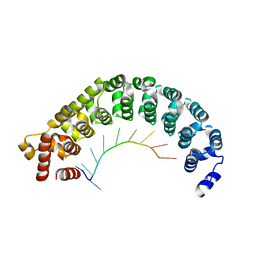 | | Puf3 RNA binding domain bound to Cox17 RNA 3' UTR recognition sequence site A | | Descriptor: | RNA (5'-R(P*CP*UP*UP*GP*UP*AP*UP*AP*UP*A)-3'), mRNA-binding protein PUF3 | | Authors: | Zhu, D, Stumpf, C.R, Krahn, J.M, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A 5' cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2BO2
 
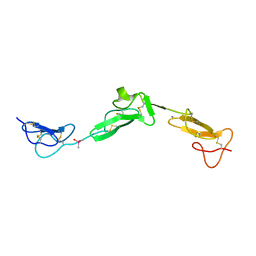 | | EGF Domains 1,2,5 of human EMR2, a 7-TM immune system molecule, in complex with calcium. | | Descriptor: | CACODYLATE ION, CALCIUM ION, EGF-LIKE MODULE CONTAINING MUCIN-LIKE HORMONE RECEPTOR-LIKE 2 PRECURSOR | | Authors: | Abbott, R.J.M, Spendlove, I, Roversi, P, Teriete, P, Knott, V, Handford, P.A, McDonnell, J.M, Lea, S.M. | | Deposit date: | 2005-04-07 | | Release date: | 2006-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Functional Characterization of a Novel T Cell Receptor Co-Regulatory Protein Complex, Cd97-Cd55.
J.Biol.Chem., 282, 2007
|
|
2BNQ
 
 | | Structural and kinetic basis for heightened immunogenicity of T cell vaccines | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, SYNTHETIC PEPTIDE, ... | | Authors: | Chen, J.-L, Stewart-Jones, G, Bossi, G, Lissin, N.M, Wooldridge, L, Choi, E.M.L, Held, G, Dunbar, P.R, Esnouf, R.M, Sami, M, Boultier, J.M, Rizkallah, P.J, Renner, C, Sewell, A, van der Merwe, P.A, Jackobsen, B.K, Griffiths, G, Jones, E.Y, Cerundolo, V. | | Deposit date: | 2005-03-31 | | Release date: | 2005-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Kinetic Basis for Heightened Immunogenicity of T Cell Vaccines
J.Exp.Med., 201, 2005
|
|
2V0Z
 
 | | Crystal Structure of Renin with Inhibitor 10 (Aliskiren) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALISKIREN, RENIN | | Authors: | Rahuel, J, Rasetti, V, Maibaum, J, Rueger, H, Goschke, R, Cohen, N.C, Stutz, S, Cumin, F, Fuhrer, W, Wood, J.M, Grutter, M.G. | | Deposit date: | 2007-05-21 | | Release date: | 2007-07-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Drug Design: The Discovery of Novel Nonpeptide Orally Active Inhibitors of Human Renin
Chem.Biol., 7, 2000
|
|
2V16
 
 | | Crystal Structure of Renin with Inhibitor 3 | | Descriptor: | METHYL (3R)-1-[(5S,6S,8R)-5-AMINO-9-BUTYLAMINO-6-HYDROXY-3,3,8-TRIMETHYL-9-OXO-NONANOYL]-3,4-DIHYDRO-2H-QUINOLINE-3-CARBOXYLATE, RENIN | | Authors: | Rahuel, J, Rasetti, V, Maibaum, J, Rueger, H, Goschke, R, Cohen, N.C, Stutz, S, Cumin, F, Fuhrer, W, Wood, J.M, Grutter, M.G. | | Deposit date: | 2007-05-22 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Drug Design: The Discovery of Novel Nonpeptide Orally Active Inhibitors of Human Renin
Chem.Biol., 7, 2000
|
|
