7PUL
 
 | | Crystal structure of Endoglycosidase E GH20 domain from Enterococcus faecalis | | Descriptor: | Beta-N-acetylhexosaminidase, CALCIUM ION, GLY-ALA-GLY-ALA-ALA, ... | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
7PUJ
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis | | Descriptor: | Beta-N-acetylhexosaminidase, CHLORIDE ION, ZINC ION | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
8EXI
 
 | |
8EXJ
 
 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain in complex with a JAK1 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Tyrosine-protein kinase JAK1 activation loop peptide, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
3QX1
 
 | | Crystal structure of FAF1 UBX domain | | Descriptor: | FAS-associated factor 1, SULFATE ION | | Authors: | Park, J.K, Jeon, H, Lee, J.J, Kim, K.H, Lee, K.J, Kim, E.E. | | Deposit date: | 2011-03-01 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissection of the interaction between FAF1 UBX and p97
To be Published
|
|
8EXM
 
 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain with a JAK3 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Tyrosine-protein kinase JAK3 activation loop peptide, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
4ZVW
 
 | | Structure of apo human ALDH7A1 in space group C2 | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J. | | Deposit date: | 2015-05-18 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
8EXN
 
 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain with TYK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-receptor tyrosine-protein kinase TYK2 activation loop peptide, PHOSPHATE ION, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
1A7B
 
 | |
8EYA
 
 | | Crystal structure of PTP1B D181A/Q262A/C215A phosphatase domain with a JAK2 activation loop phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-26 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
7Q1Y
 
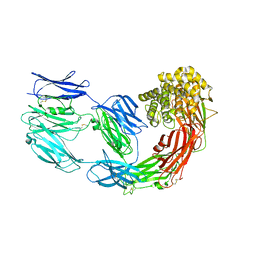 | | X-ray structure of human A2ML1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin-like protein 1, ... | | Authors: | Andersen, G.R, Zarantonello, A, Enghild, J.J, Nielsen, N.S. | | Deposit date: | 2021-10-22 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Cryo-EM structures of human A2ML1 elucidate the protease-inhibitory mechanism of the A2M family.
Nat Commun, 13, 2022
|
|
3E7X
 
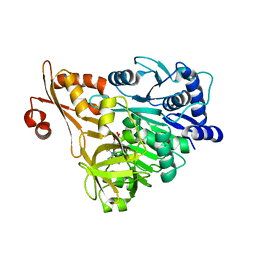 | | Crystal structure of DLTA: implications for the reaction mechanism of non-ribosomal peptide synthetase (NRPS) adenylation domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, D-alanine--poly(phosphoribitol) ligase subunit 1 | | Authors: | Yonus, H, Neumann, P, Zimmermann, S, May, J.J, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2008-08-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains
J.Biol.Chem., 283, 2008
|
|
8EYB
 
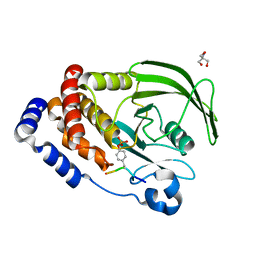 | | Crystal structure of PTP1B D181A/Q262A/C215A phosphatase domain with JAK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein kinase JAK2 activation loop phosphopeptide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-26 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
5AGR
 
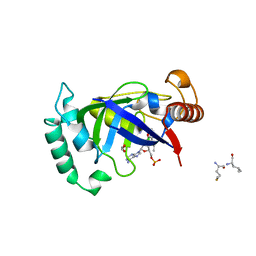 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct (S)-3-(Aminomethyl)-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 1,2-ETHANEDIOL, 3-AMINOMETHYL-7-(ETHOXY)-3H-BENZO[C][1,2]OXABOROL-1-OL modified adenosine, LEUCINE, ... | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
8EXK
 
 | |
4HPK
 
 | | Crystal structure of Clostridium histolyticum colg collagenase collagen-binding domain 3B at 1.35 Angstrom resolution in presence of calcium nitrate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collagenase, ... | | Authors: | Philominathan, S.T.L, Wilson, J.J, Bauer, R, Matsushita, O, Sakon, J. | | Deposit date: | 2012-10-24 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
7ARE
 
 | | DNA origami pointer object v2 | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Thomas, M, Feigl, E, Kohler, F, Kube, M, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
6MCC
 
 | |
3E2R
 
 | |
4HIP
 
 | |
7AS5
 
 | | 126 helix bundle DNA nanostructure | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7ART
 
 | | 48 helix bundle DNA origami brick | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Feigl, E, Kube, M, Kohler, F, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
6M7K
 
 | | Structure of mouse RECON (AKR1C13) in complex with cyclic AMP-AMP-GMP (cAAG) | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C13, cyclic AMP-AMP-GMP | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-08-20 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
4HR4
 
 | |
4HR5
 
 | | R2-like ligand-binding oxidase without metal cofactor | | Descriptor: | PALMITIC ACID, Ribonuleotide reductase small subunit | | Authors: | Griese, J.J, Hogbom, M. | | Deposit date: | 2012-10-26 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Direct observation of structurally encoded metal discrimination and ether bond formation in a heterodinuclear metalloprotein
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
