2PY7
 
 | |
9C8X
 
 | |
9C8W
 
 | |
9BYJ
 
 | | Crystal Structure of Hck in complex with the Src-family kinase inhibitor A-419259 | | Descriptor: | 1,2-ETHANEDIOL, 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DIMETHYL SULFOXIDE, ... | | Authors: | Selzer, A.M, Alvarado, J.J, Smithgall, T.E. | | Deposit date: | 2024-05-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cocrystallization of the Src-Family Kinase Hck with the ATP-Site Inhibitor A-419259 Stabilizes an Extended Activation Loop Conformation.
Biochemistry, 63, 2024
|
|
9GD1
 
 | | Structure of Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9F6C
 
 | | Cardiac myosin motor domain in the pre-powerstroke state co-crystallized with the inhibitor aficamten | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin-7, ... | | Authors: | Robert-Paganin, J, Hartman, J.J, Morgan, B.P, Malik, F.I, Houdusse, A. | | Deposit date: | 2024-05-01 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Aficamten is a small-molecule cardiac myosin inhibitor designed to treat hypertrophic cardiomyopathy.
Nat Cardiovasc Res, 3, 2024
|
|
8Y2I
 
 | |
9CE4
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 indazole inhibitor compound 6 | | Descriptor: | (1S,4r)-4-[(1P)-5-chloro-1-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-6-yl]-1-(oxetan-3-yl)piperidin-1-ium, 1,2-ETHANEDIOL, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L, Zebisch, M, Henry, C, Barker, J.J. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Discovery and Optimization of N-Heteroaryl Indazole LRRK2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
9JE1
 
 | | Human URAT1 bound to dotinurad | | Descriptor: | Solute carrier family 22 member 12, dotinurad | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
9F6D
 
 | | Human DNA polymerase epsilon bound to DNA and PCNA (open conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 2024
|
|
9JE0
 
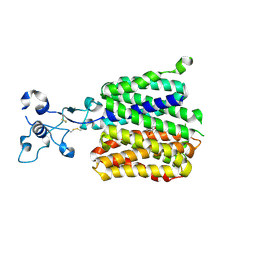 | | Human URAT1 bound to benzbromarone | | Descriptor: | Solute carrier family 22 member 12, [3,5-bis(bromanyl)-4-oxidanyl-phenyl]-(2-ethyl-1-benzofuran-3-yl)methanone | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
9LDT
 
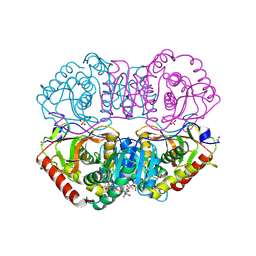 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
9F6L
 
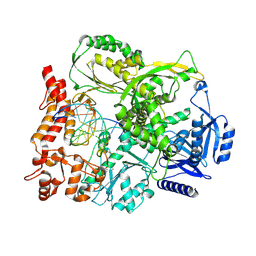 | |
9C90
 
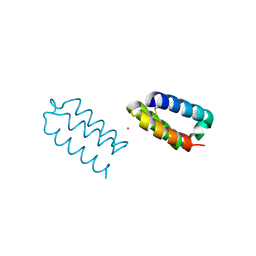 | |
9F6J
 
 | |
9GD3
 
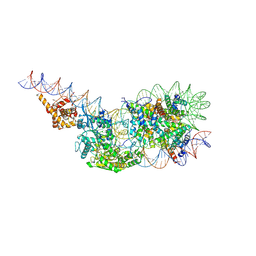 | | Structure of a mononucleosome bound by one copy of Chd1 with the DBD on the exit-side DNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9LDB
 
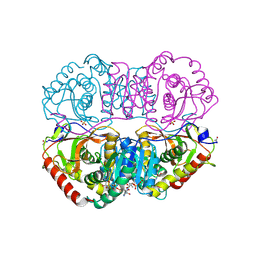 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
9C8Z
 
 | |
8XYC
 
 | |
9F6E
 
 | | Human DNA polymerase epsilon bound to DNA and PCNA (ajar conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 2024
|
|
9C8Y
 
 | |
9F6F
 
 | | Human DNA polymerase epsilon bound to DNA and PCNA (closed conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 2024
|
|
9FSE
 
 | | Human ROR2 cysteine-rich domain (CRD) and Kringle domain | | Descriptor: | SULFATE ION, Tyrosine-protein kinase transmembrane receptor ROR2 | | Authors: | Griffiths, S.C, Tan, J, Wagner, A, Blazer, L.L, Adams, J.J, Srinivasan, S, Moghisaei, S, Sidhu, S.S, Siebold, C, Ho, H.H. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and function of the ROR2 cysteine-rich domain in vertebrate noncanonical WNT5A signaling.
Elife, 13, 2024
|
|
9GD2
 
 | | Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9F6K
 
 | |
