4AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 'GREEN' SUBSTATE, AVERAGE STRUCTURE | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-29 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
2VSF
 
 | | Structure of XPD from Thermoplasma acidophilum | | Descriptor: | CALCIUM ION, DNA REPAIR HELICASE RAD3 RELATED PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Kuper, J, Wolski, S.C, Truglio, J.J, Kisker, C. | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Fes Cluster-Containing Nucleotide Excision Repair Helicase Xpd.
Plos Biol., 6, 2008
|
|
2WFG
 
 | | Structure of the Candida albicans cytosolic leucyl-tRNA synthetase editing domain bound to a benzoxaborole-AMP adduct | | Descriptor: | CYTOSOLIC LEUCYL-TRNA SYNTHETASE, [(1S,3S,5R,6R,8R)-6-(6-AMINOPURIN-9-YL)-4'-ETHYLAMINO-3'-FLUORO-SPIRO[2,4,7-TRIOXA-3-BORANUIDABICYCLO[3.3.0]OCTANE-3,7'-8-OXA-7-BORANUIDABICYCLO[4.3.0]NONA-1,3,5-TRIENE]-8-YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Seiradake, E, Mao, W, Hernandez, V, Baker, S.J, Plattner, J.J, Alley, M.R.K, Cusack, S. | | Deposit date: | 2009-04-05 | | Release date: | 2009-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of the Human and Fungal Cytosolic Leucyl-tRNA Synthetase Editing Domains: A Structural Basis for the Rational Design of Antifungal Benzoxaboroles.
J.Mol.Biol., 390, 2009
|
|
2WFE
 
 | | Structure of the Candida albicans cytosolic leucyl-tRNA synthetase editing domain | | Descriptor: | CYTOSOLIC LEUCYL-TRNA SYNTHETASE | | Authors: | Seiradake, E, Mao, W, Hernandez, V, Baker, S.J, Plattner, J.J, Alley, M.R.K, Cusack, S. | | Deposit date: | 2009-04-04 | | Release date: | 2009-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of the Human and Fungal Cytosolic Leucyl-tRNA Synthetase Editing Domains: A Structural Basis for the Rational Design of Antifungal Benzoxaboroles.
J.Mol.Biol., 390, 2009
|
|
8TCV
 
 | | Structure of PYCR1 complexed with 4-bromobenzene-1,3-dicarboxylic acid | | Descriptor: | 4-bromobenzene-1,3-dicarboxylic acid, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
8TCW
 
 | |
8TCZ
 
 | |
2W7N
 
 | | Crystal Structure of KorA Bound to Operator DNA: Insight into Repressor Cooperation in RP4 Gene Regulation | | Descriptor: | 5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*GP*CP *TP*AP*AP*AP*CP*AP*AP*G)-3', 5'-D(*CP*BRUP*BRUP*GP*TP*TP*TP*AP*GP*CP*TP*AP *AP*AP*CP*AP*BRUP*T)-3', TRFB TRANSCRIPTIONAL REPRESSOR PROTEIN | | Authors: | Koenig, B, Mueller, J.J, Lanka, E, Heinemann, U. | | Deposit date: | 2008-12-23 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Kora Bound to Operator DNA: Insight Into Repressor Cooperation in Rp4 Gene Regulation
Nucleic Acids Res., 37, 2009
|
|
8TCX
 
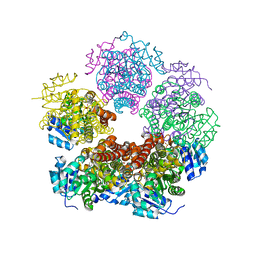 | | Structure of PYCR1 complexed with 2,4-dioxo-1,2,3,4-tetrahydroquinazoline-6-carboxylic acid | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydroquinazoline-6-carboxylic acid, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
8TD0
 
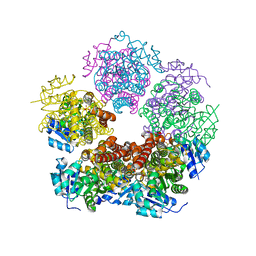 | | Structure of PYCR1 complexed with 5-oxo-7a-phenyl-hexahydropyrrolo[2,1-b][1,3]thiazole-3-carboxylic acid | | Descriptor: | (3R,4S,7aR)-5-oxo-7a-phenylhexahydropyrrolo[2,1-b][1,3]thiazole-3-carboxylic acid, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
8TCU
 
 | |
8TD1
 
 | |
8TCY
 
 | | Structure of PYCR1 complexed with 7-fluoro-2-oxo-1,2,3,4-tetrahydroquinoline-6-carboxylic acid | | Descriptor: | 7-fluoro-2-oxo-1,2,3,4-tetrahydroquinoline-6-carboxylic acid, DI(HYDROXYETHYL)ETHER, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
2VJI
 
 | | Tailspike protein of E.coli bacteriophage HK620 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, CHLORIDE ION, ... | | Authors: | Mueller, J.J, Barbirz, S, Uetrecht, C, Seckler, R, Heinemann, U. | | Deposit date: | 2007-12-11 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structure of Escherichia Coli Phage Hk620 Tailspike: Podoviral Tailspike Endoglycosidase Modules are Evolutionarily Related.
Mol.Microbiol., 69, 2008
|
|
2VFM
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666) | | Descriptor: | BIFUNCTIONAL TAIL PROTEIN, CALCIUM ION, GLYCEROL, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
1G4E
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | THIAMIN PHOSPHATE SYNTHASE | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-10-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
4JJR
 
 | |
4LH0
 
 | |
1RV5
 
 | | COMPLEX OF ECORV ENDONUCLEASE WITH D(AAAGAT)/D(ATCTT) | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*T*AP*TP*CP*TP*T)-3', ECORV ENDONUCLEASE | | Authors: | Horton, N.C, Perona, J.J. | | Deposit date: | 1998-06-01 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of protein-induced bending in the specificity of DNA recognition: crystal structure of EcoRV endonuclease complexed with d(AAAGAT) + d(ATCTT).
J.Mol.Biol., 277, 1998
|
|
2VO6
 
 | | Structure of PKA-PKB chimera complexed with 4-(4-Chlorobenzyl)-1-(7H- pyrrolo(2,3-d)pyrimidin-4-yl)piperidin-4-ylamine | | Descriptor: | 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-aminium, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Caldwell, J.J, Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Aherne, G.W, Hunter, L.K, Taylor, K, Ruddle, R, Raynaud, F.I, Verdonk, M, Workman, P, Garrett, M.D, Collins, I. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Identification of 4-(4-Aminopiperidin-1-Yl)-7H-Pyrrolo[2,3-D]Pyrimidines as Selective Inhibitors of Protein Kinase B Through Fragment Elaboration.
J.Med.Chem., 51, 2008
|
|
2VO3
 
 | | Structure of PKA-PKB chimera complexed with C-(4-(4-Chlorophenyl)-1-(7H-pyrrolo(2,3-d)pyrimidin-4-yl)piperidin-4-yl)methylamine | | Descriptor: | 1-[4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-yl]methanamine, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Caldwell, J.J, Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Aherne, G.W, Hunter, L.K, Taylor, K, Ruddle, R, Raynaud, F.I, Verdonk, M, Workman, P, Garrett, M.D, Collins, I. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Identification of 4-(4-Aminopiperidin-1-Yl)-7H-Pyrrolo[2,3-D]Pyrimidines as Selective Inhibitors of Protein Kinase B Through Fragment Elaboration.
J.Med.Chem., 51, 2008
|
|
2VNW
 
 | | Structure of PKA-PKB chimera complexed with (1-(9H-Purin-6-yl) piperidin-4-yl)methanamine | | Descriptor: | 1-[1-(9H-purin-6-yl)piperidin-4-yl]methanamine, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Caldwell, J.J, Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Aherne, G.W, Hunter, L.K, Taylor, K, Ruddle, R, Raynaud, F.I, Verdonk, M, Workman, P, Garrett, M.D, Collins, I. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Identification of 4-(4-Aminopiperidin-1-Yl)-7H-Pyrrolo[2,3-D]Pyrimidines as Selective Inhibitors of Protein Kinase B Through Fragment Elaboration.
J.Med.Chem., 51, 2008
|
|
2WZK
 
 | | Structure of the Cul5 N-terminal domain at 2.05A resolution. | | Descriptor: | 1,2-ETHANEDIOL, CULLIN-5 | | Authors: | Muniz, J.R.C, Ayinampudi, V, Zhang, Y, Babon, J.J, Chaikuad, A, Krojer, T, Pike, A.C.W, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bullock, A.N. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Architecture of the Ankyrin Socs Box Family of Cul5-Dependent E3 Ubiquitin Ligases
J.Mol.Biol., 425, 2013
|
|
1RH4
 
 | | RH4 DESIGNED RIGHT-HANDED COILED COIL TETRAMER | | Descriptor: | ISOPROPYL ALCOHOL, RIGHT-HANDED COILED COIL TETRAMER | | Authors: | Harbury, P.B, Plecs, J.J, Tidor, B, Alber, T, Kim, P.S. | | Deposit date: | 1998-10-07 | | Release date: | 1998-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein design with backbone freedom.
Science, 282, 1998
|
|
4LH3
 
 | |
