6OFE
 
 | |
6OFG
 
 | | In vitro polymerized PrgI V67A filaments | | Descriptor: | Protein PrgI | | Authors: | Guo, E.Z, Galan, J.E. | | Deposit date: | 2019-03-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A polymorphic helix of a Salmonella needle protein relays signals defining distinct steps in type III secretion.
Plos Biol., 17, 2019
|
|
1M3V
 
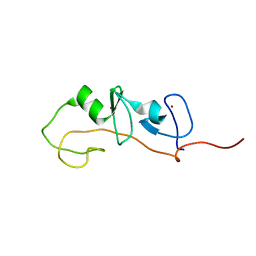 | | FLIN4: Fusion of the LIM binding domain of Ldb1 and the N-terminal LIM domain of LMO4 | | Descriptor: | ZINC ION, fusion of the LIM interacting domain of ldb1 and the N-terminal LIM domain of LMO4 | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H.Y, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2002-06-30 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
6OL8
 
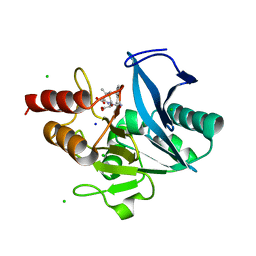 | | Crystal structure of NDM-12 metallo-beta-lactamase in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-12, ... | | Authors: | Raczynska, J.E, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
6OFH
 
 | |
6OAC
 
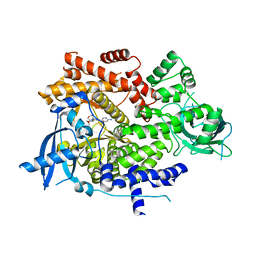 | | PQR530 [(S)-4-(Difluoromethyl)-5-(4-(3-methylmorpholino)-6-morpholino-1,3,5-triazin-2-yl)pyridin-2-amine] bound to the PI3Ka catalytic subunit p110alpha | | Descriptor: | 4-(difluoromethyl)-5-{4-[(3S)-3-methylmorpholin-4-yl]-6-(morpholin-4-yl)-1,3,5-triazin-2-yl}pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Burke, J.E, McPhail, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | (S)-4-(Difluoromethyl)-5-(4-(3-methylmorpholino)-6-morpholino-1,3,5-triazin-2-yl)pyridin-2-amine (PQR530), a Potent, Orally Bioavailable, and Brain-Penetrable Dual Inhibitor of Class I PI3K and mTOR Kinase.
J.Med.Chem., 62, 2019
|
|
6OGO
 
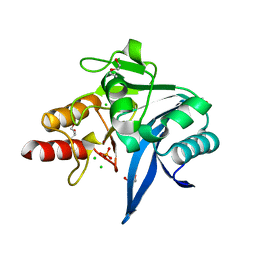 | | Crystal structure of NDM-9 metallo-beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Raczynska, J.E, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
2CK3
 
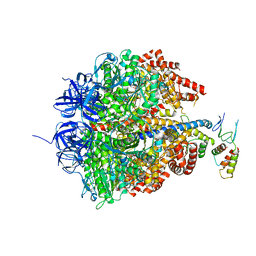 | | Azide inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE SUBUNIT ALPHA, MITOCHONDRIAL, ... | | Authors: | Bowler, M.W, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How Azide Inhibits ATP Hydrolysis by the F-Atpases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
8QH6
 
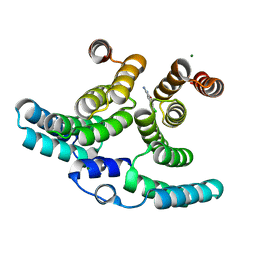 | | Crystal structure of IpgC in complex with a follow-up compound based on J20 | | Descriptor: | 3-azanyl-6-chloranyl-isoindol-1-one, CHLORIDE ION, Chaperone protein IpgC, ... | | Authors: | Wallbaum, J.E, Heine, A, Reuter, K. | | Deposit date: | 2023-09-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Fragment Screening on the Shigella Type III Secretion System Chaperone IpgC.
Acs Omega, 8, 2023
|
|
3AT1
 
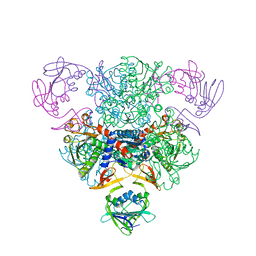 | |
3C8E
 
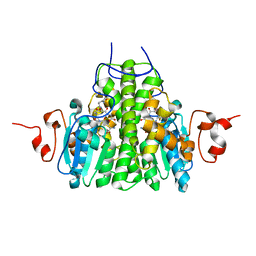 | | Crystal Structure Analysis of yghU from E. Coli | | Descriptor: | GLUTATHIONE, yghU, glutathione S-transferase homologue | | Authors: | Harp, J, Ladner, J.E, Schaab, M.R, Stourman, N.V, Armstrong, R.N. | | Deposit date: | 2008-02-11 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Function of YghU, a Nu-Class Glutathione Transferase Related to YfcG from Escherichia coli.
Biochemistry, 50, 2011
|
|
3CG5
 
 | |
3BZN
 
 | | Crystal Structure of Open form of Menaquinone-Specific Isochorismate Synthase, MenF | | Descriptor: | MAGNESIUM ION, Menaquinone-specific isochorismate synthase, SULFATE ION | | Authors: | Parsons, J.F, Shi, K.M, Ladner, J.E. | | Deposit date: | 2008-01-18 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of isochorismate synthase in complex with magnesium.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3C8P
 
 | | X-ray structure of Viscotoxin A1 from Viscum album L. | | Descriptor: | Viscotoxin A1 | | Authors: | Pal, A, Debreczeni, J.E, Sevvana, M, Gruene, T, Kahle, B, Zeeck, A, Sheldrick, G.M. | | Deposit date: | 2008-02-13 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of viscotoxins A1 and B2 from European mistletoe solved using native data alone
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3D7J
 
 | | SCO6650, a 6-pyruvoyltetrahydropterin synthase homolog from Streptomyces coelicolor | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein SCO6650 | | Authors: | Spoonamore, J.E, Roberts, S.A, Heroux, A, Bandarian, V. | | Deposit date: | 2008-05-21 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a 6-pyruvoyltetrahydropterin synthase homolog from Streptomyces coelicolor.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3CP6
 
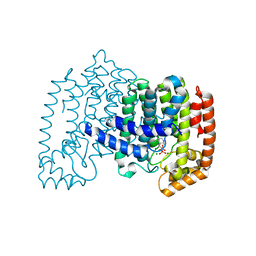 | | Crystal structure of human farnesyl diphosphate synthase (T201A mutant) complexed with Mg and biphosphonate inhibitor | | Descriptor: | (4aS,7aR)-octahydro-1H-cyclopenta[b]pyridine-6,6-diylbis(phosphonic acid), Farnesyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Pilka, E.S, Dunford, J.E, Guo, K, Pike, A.C.W, von Delft, F, Barnett, B.L, Ebetino, F.H, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Russell, R.G.G, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human farnesyl diphosphate synthase (T201A mutant) complexed with Mg and biphosphonate inhibitor.
To be Published
|
|
3DGG
 
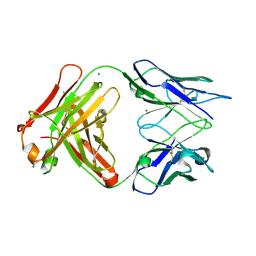 | | Crystal structure of FabOX108 | | Descriptor: | FabOX108 Heavy Chain Fragment, FabOX108 Light Chain Fragment, MAGNESIUM ION | | Authors: | Ren, J, Nettleship, J.E, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2008-06-13 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A pipeline for the production of antibody fragments for structural studies using transient expression in HEK 293T cells.
Protein Expr.Purif., 62, 2008
|
|
3DIF
 
 | |
3CSY
 
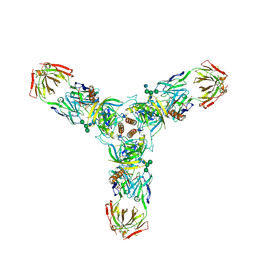 | | Crystal structure of the trimeric prefusion Ebola virus glycoprotein in complex with a neutralizing antibody from a human survivor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein GP1, ... | | Authors: | Lee, J.E, Fusco, M.L, Hessell, A.J, Oswald, W.B, Burton, D.R, Saphire, E.O. | | Deposit date: | 2008-04-10 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor.
Nature, 454, 2008
|
|
3CLC
 
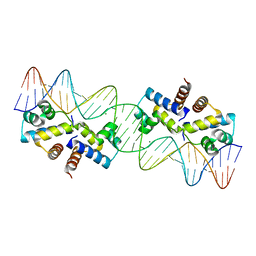 | | Crystal Structure of the Restriction-Modification Controller Protein C.Esp1396I Tetramer in Complex with its Natural 35 Base-Pair Operator | | Descriptor: | 35-MER, MAGNESIUM ION, Regulatory protein | | Authors: | McGeehan, J.E, Streeter, S.D, Thresh, S.J, Ball, N, Ravelli, R.B, Kneale, G.G. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of the genetic switch that regulates the expression of restriction-modification genes.
Nucleic Acids Res., 36, 2008
|
|
3EZR
 
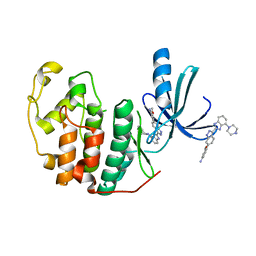 | | CDK-2 with indazole inhibitor 17 bound at its active site | | Descriptor: | 3-methoxy-4-{3-[4-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2 | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-10-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3CR1
 
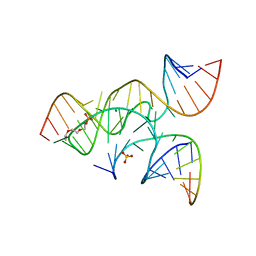 | | crystal structure of a minimal, mutant, all-RNA hairpin ribozyme (A38C, A-1OMA) grown from MgCl2 | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3'), RNA (5'-R(*UP*CP*GP*UP*GP*GP*UP*CP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3'), ... | | Authors: | Salter, J.D, Wedekind, J.E. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3DOP
 
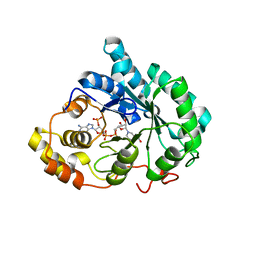 | | Crystal structure of 5beta-reductase (AKR1D1) in complex with NADP+ and 5beta-dihydrotestosterone, Resolution 2.00A | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, 5-beta-DIHYDROTESTOSTERONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Di Costanzo, L, Drury, J.E, Penning, T.M, Christianson, D.W. | | Deposit date: | 2008-07-05 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and catalytic mechanism of human steroid 5beta-reductase (AKR1D1)
Mol.Cell.Endocrinol., 301, 2009
|
|
3CQS
 
 | | A 3'-OH, 2',5'-phosphodiester substitution in the hairpin ribozyme active site reveals similarities with protein ribonucleases | | Descriptor: | 13-mer substrate strand with 3'-OH, 2',5'-phosphodiester covalently linking 5th and 6th nucleotides, 19-mer ribozyme strand, ... | | Authors: | Torelli, A.T, Spitale, R.C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Shared traits on the reaction coordinates of ribonuclease and an RNA enzyme
Biochem.Biophys.Res.Commun., 371, 2008
|
|
3F5X
 
 | | CDK-2-Cyclin complex with indazole inhibitor 9 bound at its active site | | Descriptor: | 4-{3-[7-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-11-04 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
