4BX9
 
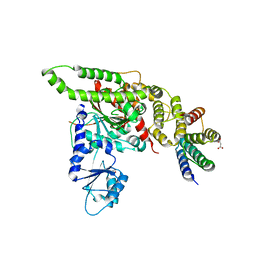 | | Human Vps33A in complex with a fragment of human Vps16 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, FORMIC ACID, MALONIC ACID, ... | | Authors: | Graham, S.C, Wartosch, L, Gray, S.R, Scourfield, E.J, Deane, J.E, Luzio, J.P, Owen, D.J. | | Deposit date: | 2013-07-09 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of Vps33A recruitment to the human HOPS complex by Vps16.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
5MN2
 
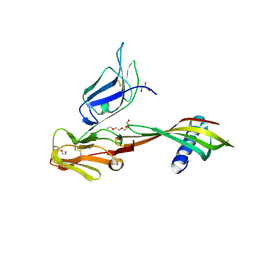 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer G3, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer G3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Robinson, J.I, Owen, R.L, Tomlinson, D.C, Baxter, E.W, Nettleship, J.E, Waterhouse, M.P, Harris, S.A, Owens, R.J, McPherson, M.J, Morgan, A.W, Tiede, C, Goldman, A, Thomsen, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EFK
 
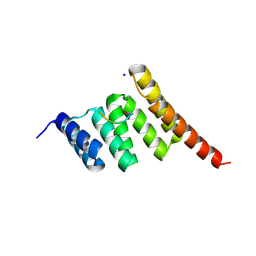 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated HSP70 peptide | | Descriptor: | ACE-ILE-GLU-GLU-VAL-ASP, E3 ubiquitin-protein ligase CHIP, SODIUM ION | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2018-08-16 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
8DQY
 
 | |
8DRI
 
 | |
4CCE
 
 | | STRUCTURE OF MOUSE GALACTOCEREBROSIDASE WITH GALACTOSE: ENZYME- PRODUCT COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Hill, C.H, Graham, S.C, Read, R.J, Deane, J.E. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Snapshots Illustrate the Catalytic Cycle of Beta-Galactocerebrosidase, the Defective Enzyme in Krabbe Disease
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8DZD
 
 | |
5KC6
 
 | |
6EQE
 
 | | High resolution crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SODIUM ION | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3GLU
 
 | | Crystal Structure of Human SIRT3 with AceCS2 peptide | | Descriptor: | Acetyl-coenzyme A synthetase 2-like, mitochondrial, NAD-dependent deacetylase sirtuin-3, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
2Z6K
 
 | | Crystal structure of full-length human RPA14/32 heterodimer | | Descriptor: | Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit | | Authors: | Deng, X, Habel, J.E, Kabaleeswaran, V, Borgstahl, G.E. | | Deposit date: | 2007-08-03 | | Release date: | 2007-12-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Full-length Human RPA14/32 Complex Gives Insights into the Mechanism of DNA Binding and Complex Formation
J.Mol.Biol., 374, 2007
|
|
1MTC
 
 | | GLUTATHIONE TRANSFERASE MUTANT Y115F | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, Glutathione S-transferase YB1 | | Authors: | Ladner, J.E, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 2002-09-20 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Local protein dynamics and catalysis: detection of segmental motion associated with rate-limiting product release by a glutathione transferase
Biochemistry, 41, 2002
|
|
1N72
 
 | | Structure and Ligand of a Histone Acetyltransferase Bromodomain | | Descriptor: | HISTONE ACETYLTRANSFERASE | | Authors: | Dhalluin, C, Carlson, J.E, Zeng, L, He, C, Aggarwal, A.K, Zhou, M.-M. | | Deposit date: | 2002-11-12 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Ligand of a Histone Acetyltransferase Bromodomain
Nature, 399, 1999
|
|
3GS5
 
 | | An all-RNA hairpin ribozyme A38N1dA variant with a product mimic substrate strand | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, COBALT HEXAMMINE(III), RNA (25-MER), ... | | Authors: | Spitale, R.C, Volpini, R, Heller, M.G, Krucinska, J, Cristalli, G, Wedekind, J.E. | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of an imino group indispensable for cleavage by a small ribozyme.
J.Am.Chem.Soc., 131, 2009
|
|
2Z8A
 
 | | Ligand Migration and Binding in The Dimeric Hemoglobin of Scapharca Inaequivalvis: I25W with CO Bound to HEME and in the Presence of 3 Atoms of XE | | Descriptor: | CARBON MONOXIDE, Globin-1, PHOSPHATE ION, ... | | Authors: | Knapp, J.E, Royer Jr, W.E, Nienhaus, K, Palladino, P, Nienhaus, G.U. | | Deposit date: | 2007-09-04 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Ligand Migration and Binding in the Dimeric Hemoglobin of Scapharca inaequivalvis
Biochemistry, 46, 2007
|
|
8EFF
 
 | | CryoEM of the soluble OPA1 tetramer from the GDP-AlFx bound helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-08 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (5.48 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EEW
 
 | | CryoEM of the soluble OPA1 dimer from the GDP-AlFx bound helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (5.48 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EFS
 
 | | CryoEM of the soluble OPA1 tetramer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EFR
 
 | | CryoEM of the soluble OPA1 interfaces with GDP-AlFx bound from the helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.48 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EF7
 
 | | CryoEM of the soluble OPA1 dimer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-08 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EFT
 
 | | CryoEM of the soluble OPA1 interfaces from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
1NWN
 
 | |
6EQF
 
 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup P212121 | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7S0N
 
 | | Structure of MS3494 from Mycobacterium Smegmatis determined by Solution NMR | | Descriptor: | Secreted protein | | Authors: | Kent, J.E, Tian, Y, Shin, K, Zhang, L, Niederweis, M, Marassi, F.M. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of MS3494 from Mycobacterium Smegmatis
To Be Published
|
|
1O99
 
 | | CRYSTAL STRUCTURE OF THE S62A MUTANT OF PHOSPHOGLYCERATE MUTASE FROM BACILLUS STEAROTHERMOPHILUS COMPLEXED WITH 2-PHOSPHOGLYCERATE | | Descriptor: | 2,3-BISPHOSPHOGLYCERATE-INDEPENDENT PHOSPHOGLYCERATE MUTASE, 2-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, ... | | Authors: | Rigden, D.J, Lamani, E, Littlejohn, J.E, Jedrzejas, M.J. | | Deposit date: | 2002-12-11 | | Release date: | 2002-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Insights Into the Catalytic Mechanism of Cofactor-Independent Phosphoglycerate Mutase from X-Ray Crystallography, Simulated Dynamics and Molecular Modeling
J.Mol.Biol., 328, 2003
|
|
