2GP1
 
 | |
6HB8
 
 | | Crystal structure of OXA-517 beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, Beta-lactamase, ... | | Authors: | Raczynska, J.E, Dabos, L, Zavala, A, Retailleau, P, Iorga, B, Jaskolski, M, Naas, T. | | Deposit date: | 2018-08-09 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Genetic, biochemical and structural characterization of OXA-517, an OXA-48-like extended-spectrum cephalosporins and carbapenems-hydrolyzing beta-lactamase
To Be Published
|
|
5N0H
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed meropenem - new refinement | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
1EBG
 
 | |
1EYF
 
 | | REFINED STRUCTURE OF THE DNA METHYL PHOSPHOTRIESTER REPAIR DOMAIN OF E. COLI ADA | | Descriptor: | ADA REGULATORY PROTEIN, ZINC ION | | Authors: | Lin, Y, Dotsch, V, Wintner, T, Peariso, K, Myers, L.C, Penner-Hahn, J.E, Verdine, G.L, Wagner, G. | | Deposit date: | 2000-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the functional switch of the E. coli Ada protein
Biochemistry, 40, 2001
|
|
2JC6
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D | | Descriptor: | CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE 1D, N-(5-METHYL-1H-PYRAZOL-3-YL)-2-PHENYLQUINAZOLIN-4-AMINE | | Authors: | Debreczeni, J.E, Rellos, P, Fedorov, O, Niesen, F.H, Bhatia, C, Shrestha, L, Salah, E, Smee, C, Colebrook, S, Berridge, G, Gileadi, O, Bunkoczi, G, Ugochukwu, E, Pike, A.C.W, von Delft, F, Knapp, S, Sundstrom, M, Weigelt, J, Arrowsmith, C.H, Edwards, A. | | Deposit date: | 2006-12-19 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Calmodulin-Dependent Protein Kinase 1D
To be Published
|
|
6HQL
 
 | | Crystal structure of GcoA F169H bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQT
 
 | | Crystal structure of GcoA F169V bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2JDI
 
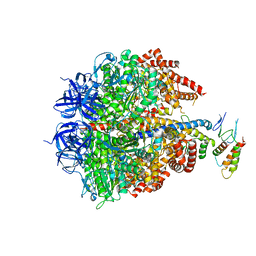 | | Ground state structure of F1-ATPase from bovine heart mitochondria (Bovine F1-ATPase crystallised in the absence of azide) | | Descriptor: | ATP SYNTHASE DELTA CHAIN, ATP SYNTHASE EPSILON CHAIN, ATP SYNTHASE GAMMA CHAIN, ... | | Authors: | Bowler, M.W, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-01-09 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ground State Structure of F1-ATPase from Bovine Heart Mitochondria at 1.9 A Resolution
J.Biol.Chem., 282, 2007
|
|
1GBI
 
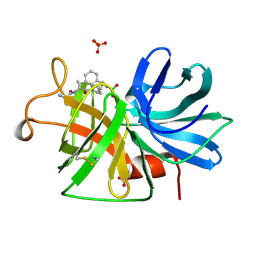 | |
1GBJ
 
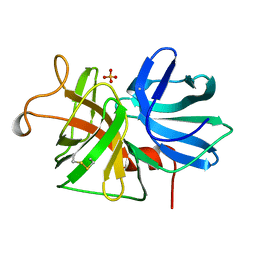 | | ALPHA-LYTIC PROTEASE WITH MET 190 REPLACED BY ALA | | Descriptor: | ALPHA-LYTIC PROTEASE, SULFATE ION | | Authors: | Mace, J.E, Agard, D.A. | | Deposit date: | 1995-09-06 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural characterization of mutations of glycine 216 in alpha-lytic protease: a new target for engineering substrate specificity.
J.Mol.Biol., 254, 1995
|
|
1GBL
 
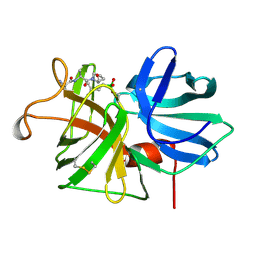 | |
1GBC
 
 | |
1GBH
 
 | |
1GBE
 
 | |
2JAM
 
 | | Crystal structure of human calmodulin-dependent protein kinase I G | | Descriptor: | 1,2-ETHANEDIOL, 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, CALCIUM ION, ... | | Authors: | Debreczeni, J.E, Bullock, A, Keates, T, Niesen, F.H, Salah, E, Shrestha, L, Smee, C, Sobott, F, Pike, A.C.W, Bunkoczi, G, von Delft, F, Turnbull, A, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Knapp, S. | | Deposit date: | 2006-11-29 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Calmodulin-Dependent Protein Kinase I G
To be Published
|
|
2JJ2
 
 | | The Structure of F1-ATPase inhibited by quercetin. | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4PXT
 
 | |
2JNC
 
 | | Refined 3D NMR structure of ECD1 of mCRF-R2beta at pH 5 | | Descriptor: | Corticotropin-releasing factor receptor 2 | | Authors: | Grace, C.R.R, Perrin, M.H, Jozsef, G, DiGruccio, M.R, Cantle, J.P, Rivier, J.E, Vale, W.W, Riek, R. | | Deposit date: | 2007-01-08 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JND
 
 | | 3D NMR structure of ECD1 of mCRF-R2b in complex with Astressin | | Descriptor: | ASTRESSIN, Corticotropin-releasing factor receptor 2 | | Authors: | Grace, C.R.R, Perrin, M.H, Jozsef, G, DiGruccio, M.R, Cantle, J.P, Rivier, J.E, Vale, W.W, Riek, R. | | Deposit date: | 2007-01-08 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JMX
 
 | | OSCP-NT (1-120) in complex with N-terminal (1-25) alpha subunit from F1-ATPase | | Descriptor: | ATP synthase O subunit, mitochondrial, ATP synthase subunit alpha heart isoform | | Authors: | Carbajo, R.J, Neuhaus, D, Kellas, F.A, Yang, J, Runswick, M.J, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2006-12-12 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | How the N-terminal Domain of the OSCP Subunit of Bovine F(1)F(o)-ATP Synthase Interacts with the N-terminal Region of an Alpha Subunit
J.Mol.Biol., 368, 2007
|
|
4RZD
 
 | | Crystal Structure of a PreQ1 Riboswitch | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, PreQ1-III Riboswitch (Class 3) | | Authors: | Wedekind, J.E, Liberman, J.A, Salim, M. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4S3M
 
 | |
4RW2
 
 | | Hen egg-white lysozyme structure from a spent-beam experiment at LCLS: refocused beam | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Foucar, L, Barends, T, Doak, R.B, Koglin, J.E, Messerschmidt, M, Nass, K, Schlichting, I, Shoeman, R, Williams, G.J. | | Deposit date: | 2014-12-01 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and use of the spent beam for serial operation of LCLS.
J.SYNCHROTRON RADIAT., 22, 2015
|
|
1GBK
 
 | |
