1E5O
 
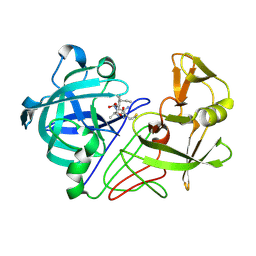 | | Endothiapepsin complex with inhibitor DB2 | | Descriptor: | ENDOTHIAPEPSIN, N-[(2S)-2-amino-3-phenylpropyl]-D-methionyl-L-alanyl-L-isoleucine | | Authors: | Read, J.A, Cooper, J.B, Toldo, L, Bailey, D. | | Deposit date: | 2000-07-28 | | Release date: | 2000-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Refinement of Four Endothiapepsin Inhibitor Complexes. Crystallographic Studies of Cytochrome Ch from Methylobacterium Extorquens and Inhibitor Complexes of Aspartic Proteinases.
Thesis, 1999
|
|
1E81
 
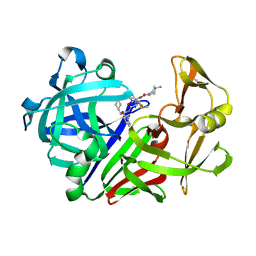 | | Endothiapepsin complex with renin inhibitor MERCK-KGAA-EMD61395 | | Descriptor: | (2S)-1-{[(2R)-1-{[(2S,3S)-1-cyclohexyl-3-hydroxy-4-(2-oxopyridin-1(2H)-yl)butan-2-yl]amino}-3-(methylsulfanyl)-1-oxopropan-2-yl]amino}-1-oxo-3-phenylpropan-2-yl 4-aminopiperidine-1-carboxylate, ENDOTHIAPEPSIN | | Authors: | Read, J.A, Cooper, J.B, Toldo, L, Rippmann, F, Raddatz, P. | | Deposit date: | 2000-09-15 | | Release date: | 2000-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Refinement of Four Endothiapepsin Inhibitor Complexes. Crystallographic Studies of Cytochrome Ch from Methylobacterium Extorquens and Inhibitor Complexes of Aspartic Proteinases.
Thesis, 1999
|
|
2B42
 
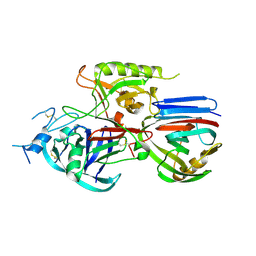 | | Crystal structure of the Triticum xylanse inhibitor-I in complex with bacillus subtilis xylanase | | Descriptor: | Endo-1,4-beta-xylanase A, xylanase inhibitor-I | | Authors: | Sansen, S, Dewilde, M, De Ranter, C.J, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-09-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of structural determinants for inhibition strength and specificity of wheat xylanase inhibitors TAXI-IA and TAXI-IIA.
Febs J., 276, 2009
|
|
1E6W
 
 | |
2AZM
 
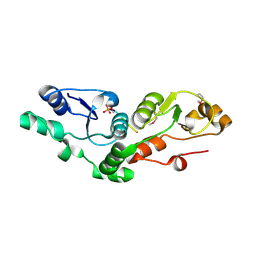 | | Crystal structure of the MDC1 brct repeat in complex with the histone tail of gamma-H2AX | | Descriptor: | GAMMA-H2AX HISTONE, Mediator of DNA damage checkpoint protein 1 | | Authors: | Clapperton, J.A, Stucki, M, Mohammad, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2005-09-12 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | MDC1 Directly Binds Phosphorylated Histone H2AX to Regulate Cellular Responses to DNA Double-Strand Breaks
Cell(Cambridge,Mass.), 123, 2005
|
|
2ATS
 
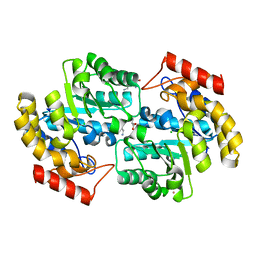 | | Dihydrodipicolinate synthase co-crystallised with (S)-lysine | | Descriptor: | CHLORIDE ION, D-LYSINE, POTASSIUM ION, ... | | Authors: | Devenish, S.R.A, Dobson, R.C.J, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-08-26 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The co-crystallisation of (S)-lysine-bound dihydrodipicolinate synthase from E. coli indicates that domain movements are not responsible for (S)-lysine inhibition
To be published
|
|
4KC2
 
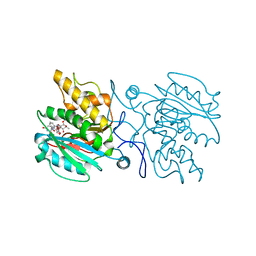 | | Structure of the blood group glycosyltransferase AAglyB in complex with a pyridine inhibitor as a neutral pyrophosphate surrogate | | Descriptor: | 6-(1-beta-D-Galactopyranosyloxymethyl)-N-(5'-deoxyluridine-5'-yl)picolinamide, Fucosylglycoprotein alpha-N-acetylgalactosaminyltransferase soluble form, MANGANESE (II) ION, ... | | Authors: | Cuesta-Seijo, J.A, Wang, S, Lafont, D, Vidal, S, Palcic, M.M. | | Deposit date: | 2013-04-24 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of glycosyltransferase inhibitors: pyridine as a pyrophosphate surrogate.
Chemistry, 19, 2013
|
|
4K5X
 
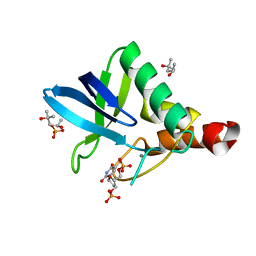 | | Crystal structure of Staphylococcal nuclease variant V23M/L36F at cryogenic temperature | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Caro, J.A, Flores, E, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-04-15 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pressure effects on proteins
To be Published
|
|
4KJN
 
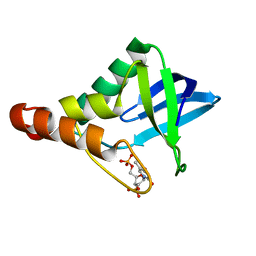 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23T/V66A/V99T at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-05-03 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cavities in proteins
To be Published
|
|
4KDU
 
 | | Crystal structure of a glutathione transferase family member from Burkholderia graminis, target efi-507264, no gsh, ordered domains, space group P21, form(1) | | Descriptor: | Glutathione S-transferase domain | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Burkholderia graminis, target efi-507264, no gsh, ordered domains, space group P21, form(1)
To be Published
|
|
4KD4
 
 | | Crystal structure of Staphylococcal nuclease variant V23L/L25V/V66I/I72V at cryogenic temperature | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Caro, J.A, Flores, E, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4KAE
 
 | | Crystal structure of Maleylacetoacetate isomerase from Anaeromyxobacter dehalogenans 2CP-1, TARGET EFI-507175, with bound dicarboxyethyl glutathione and citrate in the active site | | Descriptor: | CITRIC ACID, GAMMA-GLUTAMYL-S-(1,2-DICARBOXYETHYL)CYSTEINYLGLYCINE, GLYCEROL, ... | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Maleylacetoacetate isomerase from Anaeromyxobacter dehalogenans 2CP-1, TARGET EFI-507175, with bound dicarboxyethyl glutathione and citrate in the active site
To be Published
|
|
4KD3
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25A/V66A/I92A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4KM5
 
 | | X-ray crystal structure of human cyclic GMP-AMP synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Berger, J.M, Doudna, J.A. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity.
Cell Rep, 3, 2013
|
|
4KPL
 
 | | Crystal structure of d-mannonate dehydratase from chromohalobacter salexigens complexed with Mg,d-mannonate and 2-keto-3-deoxy-d-gluconate | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, CHLORIDE ION, D-MANNONIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-05-13 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of d-mannonate dehydratase from chromohalobacter salexigens complexed with Mg,d-mannonate and 2-keto-3-deoxy-d-gluconate
To be Published
|
|
4KQW
 
 | | The structure of the Slackia exigua KARI in complex with NADP | | Descriptor: | Ketol-acid reductoisomerase, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Brinkmann-Chen, S, Flock, T, Cahn, J.K.B, Snow, C.D, Brustad, E.M, Mcintosh, J.A, Meinhold, P, Zhang, L, Arnold, F.H. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KGI
 
 | | Crystal structure of a glutathione transferase family member from Shigella flexneri, target EFI-507258, bound GSH, TEV-his-tag linker in active site | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Shigella flexneri, target EFI-507258, bound GSH, TEV-His-tag linker in active site
To be published
|
|
4KVY
 
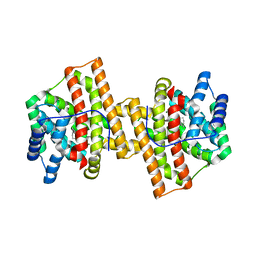 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with (1S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | Descriptor: | (1S,5S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Al-lami, N, Janvier, M, D'Antonio, E.L, Faraldos, J.A, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
4KUX
 
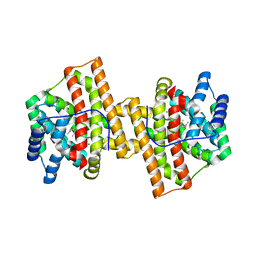 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with farnesyl thiolodiphosphate (FSPP) | | Descriptor: | Aristolochene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, M, Faraldos, J.A, Al-lami, N, Janvier, M, D'Antonio, E.L, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
4L0N
 
 | | Crystal structure of STK3 (MST2) SARAH domain | | Descriptor: | Serine/threonine-protein kinase 3 | | Authors: | Chaikuad, A, Krojer, T, Newman, J.A, Dixon-Clarke, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-05-31 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of STK3 (MST2) SARAH domain
To be Published
|
|
5TCA
 
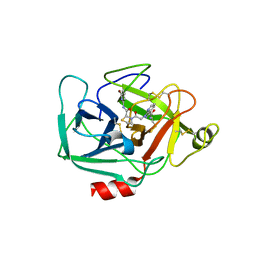 | | Complement Factor D inhibited with JH3 | | Descriptor: | 1-(2-{(2S)-2-[(6-bromopyridin-2-yl)carbamoyl]-1,3-thiazolidin-3-yl}-2-oxoethyl)-1H-pyrazolo[3,4-b]pyridine-3-carboxamide, Complement factor D | | Authors: | Stuckey, J.A. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Buried Hydrogen Bond Interactions Contribute to the High Potency of Complement Factor D Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5TDQ
 
 | | Crystal Structure of the GOLD domain of ACBD3 | | Descriptor: | Golgi resident protein GCP60 | | Authors: | McPhail, J.A, Burke, J.E. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | The Molecular Basis of Aichi Virus 3A Protein Activation of Phosphatidylinositol 4 Kinase III beta , PI4KB, through ACBD3.
Structure, 25, 2017
|
|
5TM2
 
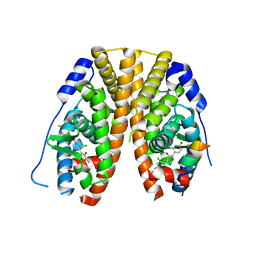 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 2,5-bis(2-chloro-4-hydroxyphenyl)thiophene 1-oxide | | Descriptor: | 2,5-bis(2-chloro-4-hydroxyphenyl)-1H-1lambda~4~-thiophen-1-one, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMS
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Cyclofenil-ASC derivative, ethyl (E)-3-(4-(bicyclo[3.3.1]nonan-9-ylidene(4-hydroxyphenyl)methyl)phenyl)acrylate | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, ethyl 3-(4-{[(1s,5s)-bicyclo[3.3.1]nonan-9-ylidene](4-hydroxyphenyl)methyl}phenyl)prop-2-enoate | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM5
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 5-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)pentanoic acid | | Descriptor: | 5-{4-[(1S,4S,5R)-5-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}pentanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
