3QJT
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3M01
 
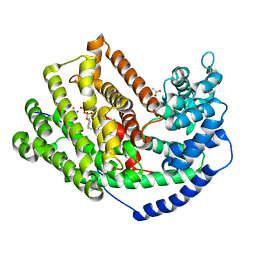 | | The Crystal Structure of 5-epi-aristolochene synthase complexed with (2-trans,6-trans)-2-fluorofarnesyl diphosphate | | Descriptor: | (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, ACETATE ION, Aristolochene synthase, ... | | Authors: | Noel, J.P, Dellas, N, Faraldos, J.A, Zhao, M, Hess Jr, B.A, Smentek, L, Coates, R.M, O'Maille, P.E. | | Deposit date: | 2010-03-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elucidation of cisoid and transoid cyclization pathways of a sesquiterpene synthase using 2-fluorofarnesyl diphosphates.
Acs Chem.Biol., 5, 2010
|
|
6ASI
 
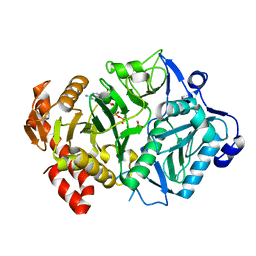 | | E. coli phosphoenolpyruvate carboxykinase G209S mutant bound to methanesulfonate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Tang, H.Y.H, Shin, D.S, Tainer, J.A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Structural Control of Nonnative Ligand Binding in Engineered Mutants of Phosphoenolpyruvate Carboxykinase.
Biochemistry, 57, 2018
|
|
6AT2
 
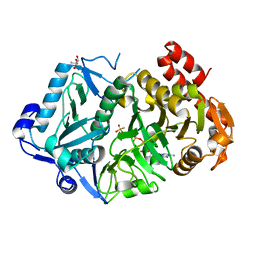 | | E. coli phosphoenolpyruvate carboxykinase G209N mutant bound to thiosulfate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tang, H.Y.H, Shin, D.S, Tainer, J.A. | | Deposit date: | 2017-08-27 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Structural Control of Nonnative Ligand Binding in Engineered Mutants of Phosphoenolpyruvate Carboxykinase.
Biochemistry, 57, 2018
|
|
3M5Z
 
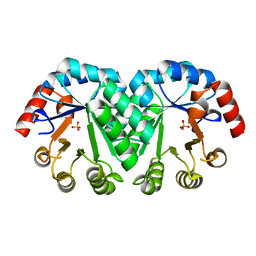 | | Crystal structure of the mutant V182A,I218A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | Orotidine 5'-phosphate decarboxylase, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
6APO
 
 | | Anti-Marburgvirus Nucleoprotein Single Domain Antibody A | | Descriptor: | Anti-Marburgvirus Nucleoprotein Single Domain Antibody A | | Authors: | Taylor, A.B, Garza, J.A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.168 Å) | | Cite: | Unveiling a Drift Resistant Cryptotope withinMarburgvirusNucleoprotein Recognized by Llama Single-Domain Antibodies.
Front Immunol, 8, 2017
|
|
4APE
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN | | Authors: | Pearl, L.H, Sewell, B.T, Jenkins, J.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1986-06-09 | | Release date: | 1986-07-14 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
6H3R
 
 | |
6GUE
 
 | | CDK2/CyclinA in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GV4
 
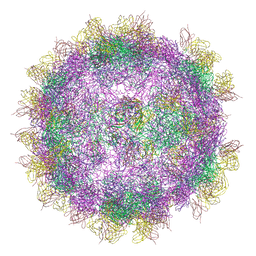 | | High-resolution Cryo-EM of Fab-labeled human parechovirus 3 | | Descriptor: | AT12-015 antibody variable heavy, AT12-015 antibody variable light, RNA (5'-R(*UP*GP*GP*UP*AP*UP*UP*U)-3'), ... | | Authors: | Domanska, A, Flatt, J.W, Jukonen, J.J.J, Geraets, J.A, Butcher, S.J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A 2.8-Angstrom-Resolution Cryo-Electron Microscopy Structure of Human Parechovirus 3 in Complex with Fab from a Neutralizing Antibody.
J.Virol., 93, 2019
|
|
3M5Y
 
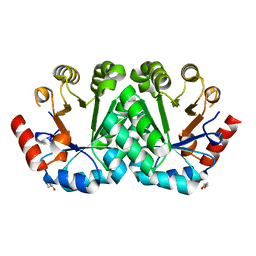 | | Crystal structure of the mutant V182A,V201A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | FORMIC ACID, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.455 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3M0T
 
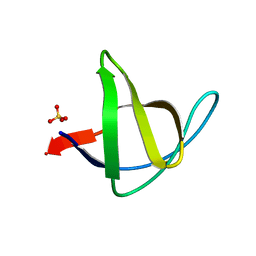 | |
6CL9
 
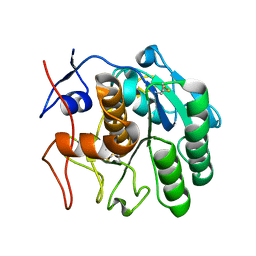 | | 2.20 A MicroED structure of proteinase K at 4.3 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.2 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLG
 
 | | 1.35 A MicroED structure of GSNQNNF at 2.4 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.35 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLN
 
 | | 1.15 A MicroED structure of GSNQNNF at 1.8 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.15 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLA
 
 | | 2.80 A MicroED structure of proteinase K at 6.0 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLJ
 
 | | 1.01 A MicroED structure of GSNQNNF at 0.50 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLQ
 
 | | 1.21 A MicroED structure of GSNQNNF at 2.8 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.21 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6E9V
 
 | | DHF79 filament | | Descriptor: | DHF79 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
3PBJ
 
 | | Hydrolytic catalysis and structural stabilization in a designed metalloprotein | | Descriptor: | CHLORIDE ION, COIL SER L9L-Pen L23H, MERCURY (II) ION, ... | | Authors: | Zastrow, M.L, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2010-10-20 | | Release date: | 2011-11-30 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hydrolytic catalysis and structural stabilization in a designed metalloprotein.
Nat Chem, 4, 2012
|
|
3PBY
 
 | | Crystal structure of the mutant L123S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
6EGN
 
 | | Crystal Structure of a Three-stranded Coiled Coil Peptide Containing a Trigonal Planar Hg(II)S3 Site Modified by D-Leu in the Second Coordination Sphere | | Descriptor: | CHLORIDE ION, Hg(II)(GRAND CoilSerL16CL19(DLE))3-, MERCURY (II) ION, ... | | Authors: | Ruckthong, L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-03 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How Outer Coordination Sphere Modifications Can Impact Metal Structures in Proteins: A Crystallographic Evaluation.
Chemistry, 25, 2019
|
|
1X6M
 
 | | Crystal structure of the glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, SULFATE ION, ... | | Authors: | Neculai, A.M, Neculai, D, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
6EHO
 
 | | Dimer of the Sortilin Vps10p domain at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thirup, S.S, Quistgaard, E.H, Januliene, D, Andersen, J.L, Nielsen, J.A. | | Deposit date: | 2017-09-14 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Acidic Environment Induces Dimerization and Ligand Binding Site Collapse in the Vps10p Domain of Sortilin.
Structure, 25, 2017
|
|
3PBW
 
 | | Crystal structure of the mutant L123N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
