6M40
 
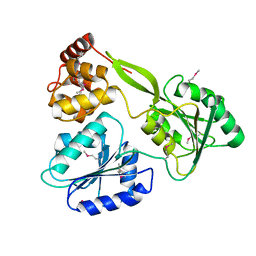 | | Crystal structure of the NS3-like helicase from Alongshan virus | | Descriptor: | NS3-like protein | | Authors: | Gao, X.P, Zhu, K.X, Chen, P, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2020-03-05 | | Release date: | 2020-04-08 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of the NS3-like helicase from Alongshan virus.
Iucrj, 7, 2020
|
|
6M7M
 
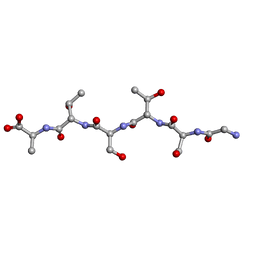 | | rac-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | L-GSTSTA from ice nucleation protein, inaZ, and its enantiomer, ... | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
6MCB
 
 | |
6M9J
 
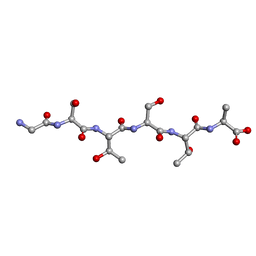 | | Racemic-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | Ice nucleation protein | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
6MMO
 
 | |
6MMQ
 
 | |
6MRH
 
 | |
6MM2
 
 | |
6MHB
 
 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 18 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutathione S-transferase omega-1, N-[4-(4-chlorophenyl)-1,3-thiazol-2-yl]propanamide | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
6MIE
 
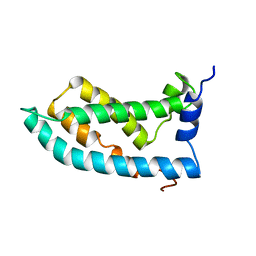 | | Solution NMR structure of the KCNQ1 voltage-sensing domain | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Taylor, K.C, Kuenze, G, Smith, J.A, Meiler, J, McFeeters, R.L, Sanders, C.R. | | Deposit date: | 2018-09-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and physiological function of the human KCNQ1 channel voltage sensor intermediate state.
Elife, 9, 2020
|
|
6MRE
 
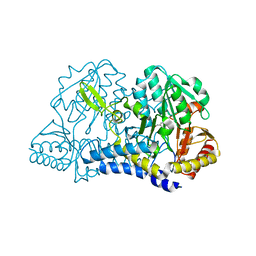 | |
6MMC
 
 | |
6MRI
 
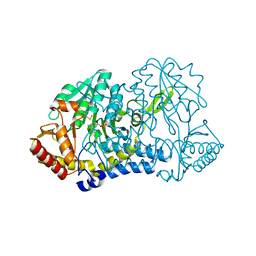 | |
6MRC
 
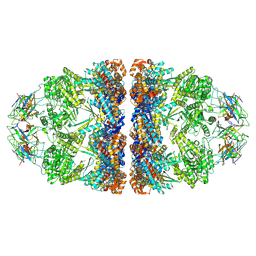 | | ADP-bound human mitochondrial Hsp60-Hsp10 football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
6MX5
 
 | |
8QIC
 
 | |
1ZZQ
 
 | | Rat nNOS D597N mutant with L-N(omega)-Nitroarginine-(4R)-amino-L-proline amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
2BV9
 
 | | HOW FAMILY 26 GLYCOSIDE HYDROLASES ORCHESTRATE CATALYSIS ON DIFFERENT POLYSACCHARIDES. STRUCTURE AND ACTIVITY OF A CLOSTRIDIUM THERMOCELLUM LICHENASE, CtLIC26A | | Descriptor: | ENDOGLUCANASE H | | Authors: | Taylor, E.J, Goyal, A, Guerreiro, C.I.P.D, Prates, J.A.M, Money, V.A, Ferry, N, Morland, C, Planas, A, Macdonald, J.A, Stick, R.V, Gilbert, H.J, Fontes, C.M.G.A, Davies, G.J. | | Deposit date: | 2005-06-23 | | Release date: | 2005-06-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How Family 26 Glycoside Hydrolases Orchestrate Catalysis on Different Polysaccharides: Structure and Activity of a Clostridium Thermocellum Lichenase, Ctlic26A.
J.Biol.Chem., 280, 2005
|
|
4DJB
 
 | | A Structural Basis for the Assembly and Functions of a Viral Polymer that Inactivates Multiple Tumor Suppressors | | Descriptor: | E4-ORF3 | | Authors: | Ou, H.D, Kwiatkowski, W, Deerinck, T.J, Noske, A, Blain, K.Y, Land, H.S, Soria, C, Powers, C.J, May, A.P, Shu, X, Tsien, R.Y, Fitzpatrick, J.A.J, Long, J.A, Ellisman, M.H, Choe, S, O'Shea, C.C. | | Deposit date: | 2012-02-01 | | Release date: | 2012-10-31 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | A Structural Basis for the Assembly and Functions of a Viral Polymer that Inactivates Multiple Tumor Suppressors.
Cell(Cambridge,Mass.), 151, 2012
|
|
2BVD
 
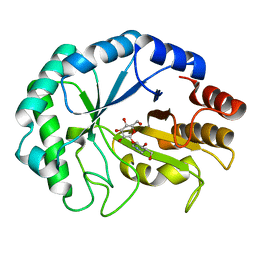 | | HOW FAMILY 26 GLYCOSIDE HYDROLASES ORCHESTRATE CATALYSIS ON DIFFERENT POLYSACCHARIDES. STRUCTURE AND ACTIVITY OF A CLOSTRIDIUM THERMOCELLUM LICHENASE, CtLIC26A | | Descriptor: | (3R,4R,5R)-4-hydroxy-5-(hydroxymethyl)piperidin-3-yl beta-D-glucopyranoside, ENDOGLUCANASE H | | Authors: | Taylor, E.J, Goyal, A, Guerreiro, C.I.P.D, Prates, J.A.M, Money, V.A, Ferry, N, Morland, C, Planas, A, Macdonald, J.A, Stick, R.V, Gilbert, H.J, Fontes, C.M.G.A, Davies, G.J. | | Deposit date: | 2005-06-27 | | Release date: | 2005-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How Family 26 Glycoside Hydrolases Orchestrate Catalysis on Different Polysaccharides: Structure and Activity of a Clostridium Thermocellum Lichenase, Ctlic26A.
J.Biol.Chem., 280, 2005
|
|
4D0R
 
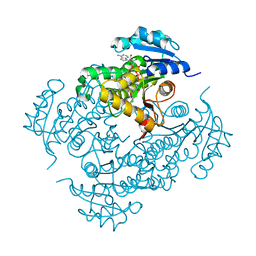 | | Mtb InhA complex with Pyradizinone compound 1 | | Descriptor: | 4-(4-chlorophenoxy)-6-oxo-1-phenyl-1,6-dihydropyridazine-3-carboxamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Read, J.A, Gingell, H, Madhavapeddi, P, Lange, S. | | Deposit date: | 2014-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Pyridazinones: A Novel Scaffold with Excellent Physicochemical Properties and Safety Profile for a Clinically Validated Target of Mycobacterium Tuberculosis
To be Published
|
|
4CK0
 
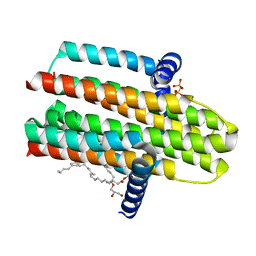 | | Crystal structure of the integral membrane diacylglycerol kinase - form 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DIACYLGLYCEROL KINASE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Li, D, Lyons, J.A, Vogeley, L, Aragao, D, Caffrey, M. | | Deposit date: | 2013-12-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.924 Å) | | Cite: | Crystal Structure of the Integral Membrane Diacylglycerol Kinase with Zn-Amppcp Bound and its Catalytic Mechanism
To be Published
|
|
4CKR
 
 | | Crystal structure of the human DDR1 kinase domain in complex with DDR1-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-ethylpiperazin-1-yl)methyl]-n-{4-methyl-3-[(2-oxo-2,3-dihydro-1h-indol-5-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Newman, J.A, Dixon-Clarke, S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective Ddr1 Receptor Tyrosine Kinase Inhibitor.
Acs Chem.Biol., 8, 2013
|
|
4D0S
 
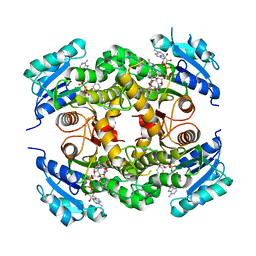 | | Mtb InhA complex with Pyradizinone compound 14 | | Descriptor: | 1-{4-[(acetylamino)methyl]phenyl}-4-(4-chlorophenoxy)-6-oxo-1,6-dihydropyridazine-3-carboxamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], MAGNESIUM ION, ... | | Authors: | Read, J.A, Breed, J, Madhavapeddi, P, Lange, S. | | Deposit date: | 2014-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Pyridazinones: A Novel Scaffold with Excellent Physicochemical Properties and Safety Profile for a Clinically Validated Target of Mycobacterium Tuberculosis
To be Published
|
|
4COQ
 
 | | The complex of alpha-Carbonic anhydrase from Thermovibrio ammonificans with inhibitor sulfanilamide. | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CARBONATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Berg, S, Lioliou, M, Kotlar, H, Littlechild, J.A. | | Deposit date: | 2014-01-30 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of a Tetrameric [Alpha]-Carbonic Anhydrase from Thermovibrio Ammonificans Reveals a Core Formed Around Intermolecular Disulfides that Contribute to its Thermostability
Acta Crystallogr.,Sect.D, 70, 2014
|
|
