2BNK
 
 | | The structure of phage phi29 replication organizer protein p16.7 | | Descriptor: | EARLY PROTEIN GP16.7 | | Authors: | Albert, A, Asensio, J.L, Munoz-Espin, D, Gonzalez, C, Hermoso, J.A, Villar, L, Jimenez-Barbero, J, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-03-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Functional Domain of {Varphi}29 Replication Organizer: Insights Into Oligomerization and DNA Binding.
J.Biol.Chem., 280, 2005
|
|
5DXM
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 3-[(E)-(1s,5s)-bicyclo[3.3.1]non-9-ylidene(4-hydroxyphenyl)methyl]phenol | | Descriptor: | 3-[(E)-(1s,5s)-bicyclo[3.3.1]non-9-ylidene(4-hydroxyphenyl)methyl]phenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5VFC
 
 | | WDR5 bound to inhibitor MM-589 | | Descriptor: | 1,2-ETHANEDIOL, N-{(3R,6S,9S,12R)-6-ethyl-12-methyl-9-[3-(N'-methylcarbamimidamido)propyl]-2,5,8,11-tetraoxo-3-phenyl-1,4,7,10-tetraazacyclotetradecan-12-yl}-2-methylpropanamide, SULFATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
7PMU
 
 | | Crystal structure of native Iripin-8 | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, Serpin-8, ... | | Authors: | Polderdijk, S, Kotal, J, Chmelar, J, Huntington, J.A. | | Deposit date: | 2021-09-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Ixodes ricinus Salivary Serpin Iripin-8 Inhibits the Intrinsic Pathway of Coagulation and Complement.
Int J Mol Sci, 22, 2021
|
|
2BW2
 
 | | BofC from Bacillus subtilis | | Descriptor: | BYPASS OF FORESPORE C | | Authors: | Patterson, H.M, Brannigan, J.A, Cutting, S.M, Wilson, K.S, Wilkinson, A.J, Ab, E, Diercks, T, Folkers, G.E, de Jong, R.N, Truffault, V, Kaptein, R. | | Deposit date: | 2005-07-08 | | Release date: | 2005-09-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of bypass of forespore C, an intercompartmental signaling factor during sporulation in Bacillus.
J. Biol. Chem., 280, 2005
|
|
6BTH
 
 | | Crystal structure of human cellular retinol binding protein 2 (CRBP2) in complex with 2-arachidonoylglycerol (2-AG) | | Descriptor: | 1,3-dihydroxypropan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, DI(HYDROXYETHYL)ETHER, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Blaner, W.S, Lodowski, D.T, Golczak, M. | | Deposit date: | 2017-12-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Retinol-binding protein 2 (RBP2) binds monoacylglycerols and modulates gut endocrine signaling and body weight.
Sci Adv, 6, 2020
|
|
5TOD
 
 | | Transmembrane protein 24 SMP domain | | Descriptor: | Transmembrane protein 24 | | Authors: | Lees, J.A, Reinisch, K.M. | | Deposit date: | 2016-10-17 | | Release date: | 2017-03-01 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion.
Science, 355, 2017
|
|
5DMC
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a nitrile-substituted triaryl-ethylene derivative 3,3-bis(4-hydroxyphenyl)-2-phenylprop-2-enenitrile | | Descriptor: | 3,3-bis(4-hydroxyphenyl)-2-phenylprop-2-enenitrile, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-08 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
2C0S
 
 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-09-07 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
6DLA
 
 | |
5TYB
 
 | | DNA Polymerase Mu Reactant Complex, 10mM Mg2+ (7.5 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-19 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
5TYV
 
 | | DNA Polymerase Mu Reactant Complex, Mn2+ (7.5 min) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
5DKB
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 3-methylphenylamino-substituted ethyl triaryl-ethylene derivative 4,4'-(2-{3-[(3-methylphenyl)amino]phenyl}but-1-ene-1,1-diyl)diphenol | | Descriptor: | 4,4'-(2-{3-[(3-methylphenyl)amino]phenyl}but-1-ene-1,1-diyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DKV
 
 | | Crystal Structure of an ABC transporter Solute Binding Protein from Agrobacterium vitis(Avis_5339, TARGET EFI-511225) bound with alpha-D-Tagatopyranose | | Descriptor: | ABC transporter substrate binding protein (Ribose), alpha-D-tagatopyranose | | Authors: | Yadava, U, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of an ABC transporter Solute Binding Protein from Agrobacterium vitis(Avis_5339, TARGET EFI-511225) bound with alpha-D-Tagatopyranose
To be published
|
|
2BJH
 
 | | Crystal Structure Of S133A AnFaeA-ferulic acid complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Faulds, C.B, Molina, R, Gonzalez, R, Husband, F, Juge, N, Sanz-Aparicio, J, Hermoso, J.A. | | Deposit date: | 2005-02-02 | | Release date: | 2005-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Probing the Determinants of Substrate Specificity of a Feruloyl Esterase, Anfaea, from Aspergillus Niger
FEBS J., 272, 2005
|
|
2BEH
 
 | | Crystal structure of antithrombin variant S137A/V317C/T401C with plasma latent antithrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III, ... | | Authors: | Johnson, D.J, Luis, S.A, Huntington, J.A. | | Deposit date: | 2005-10-24 | | Release date: | 2005-11-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation.
J.Biol.Chem., 281, 2006
|
|
2BN0
 
 | | Banana Lectin bound to Laminaribiose | | Descriptor: | CADMIUM ION, RIPENING-ASSOCIATED PROTEIN, SULFATE ION, ... | | Authors: | Meagher, J.L, Winter, H.C, Ezell, P, Goldstein, I.J, Stuckey, J.A. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Banana Lectin Reveals a Novel Second Sugar Binding Site.
Glycobiology, 15, 2005
|
|
5TYY
 
 | | DNA Polymerase Mu Product Complex, Mn2+ (60 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
5E7M
 
 | |
5DS6
 
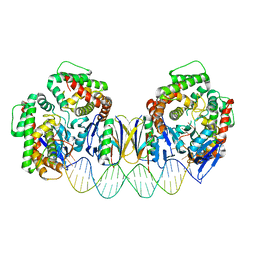 | | Crystal structure the Escherichia coli Cas1-Cas2 complex bound to protospacer DNA with splayed ends | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | Authors: | Nunez, J.K, Harrington, L.B, Kranzusch, P.J, Engelman, A.N, Doudna, J.A. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.352 Å) | | Cite: | Foreign DNA capture during CRISPR-Cas adaptive immunity.
Nature, 527, 2015
|
|
5VVJ
 
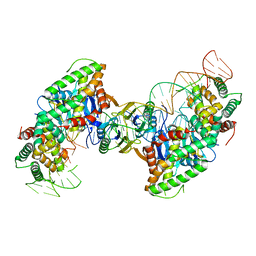 | | Cas1-Cas2 bound to half-site intermediate | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (112-MER), ... | | Authors: | Wright, A.V, Knott, G.J, Doxzen, K.W, Doudna, J.A. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
6DS9
 
 | | Elongated version of a de novo designed three helix bundle structure (GRa3D) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THIOCYANATE ION, ... | | Authors: | Koebke, K.J, Ruckthong, L.R, Meagher, J.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Clarifying the Copper Coordination Environment in a de Novo Designed Red Copper Protein.
Inorg Chem, 57, 2018
|
|
5DP0
 
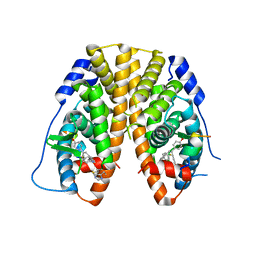 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 4-fluorophenylamino-substituted triaryl-ethylene derivative 4,4'-(2-{3-[(4-fluorophenyl)amino]phenyl}ethene-1,1-diyl)diphenol | | Descriptor: | 4,4'-(2-{3-[(4-fluorophenyl)amino]phenyl}ethene-1,1-diyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
1Z2U
 
 | | The 1.1A crystallographic structure of ubiquitin-conjugating enzyme (ubc-2) from Caenorhabditis elegans: functional and evolutionary significance | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gavira, J.A, DiGiamamarino, E, Tempel, W, Liu, Z.J, Wang, B.C, Meehan, E, Ng, J.D, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1A crystallographic structure of ubiquitin-conjugating enzyme (ubc-2) from Caenorhabditis elegans: functional and evolutionary significance
To be published
|
|
5DYB
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-(3,4-dihydronaphthalen-2(1H)-ylidenemethanediyl)diphenol | | Descriptor: | 4,4'-(3,4-dihydronaphthalen-2(1H)-ylidenemethanediyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-24 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
