3RO6
 
 | | Crystal structure of Dipeptide Epimerase from Methylococcus capsulatus complexed with Mg ion | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative chloromuconate cycloisomerase, ... | | Authors: | Lukk, T, Sakai, A, Song, L, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2011-04-25 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3R10
 
 | | Crystal structure of NYSGRC enolase target 200555, a putative dipeptide epimerase from Francisella philomiragia : Mg complex | | Descriptor: | Enzyme of enolase superfamily, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-03-09 | | Release date: | 2011-04-20 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3OTU
 
 | | PDK1 mutant bound to allosteric disulfide fragment activator JS30 | | Descriptor: | 3-(1H-INDOL-3-YL)-4-{1-[2-(1-METHYLPYRROLIDIN-2-YL)ETHYL]-1H-INDOL-3-YL}-1H-PYRROLE-2,5-DIONE, 3-phosphoinositide-dependent protein kinase 1, 4-[4-(naphthalen-1-ylmethyl)piperazin-1-yl]-4-oxobutane-1-thiol, ... | | Authors: | Sadowsky, J.D, Wells, J.A. | | Deposit date: | 2010-09-13 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1013 Å) | | Cite: | Turning a protein kinase on or off from a single allosteric site via disulfide trapping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OW1
 
 | | Crystal structure of D-mannonate dehydratase from Chromohalobacter salexigens complexed with MG | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | CRYSTAL STRUCTURE OF D-MANNONATE DEHYDRATASE FROM CHROMOHALOBACTER SALEXIGENS complexed with MG
To be Published
|
|
1Q41
 
 | | GSK-3 Beta complexed with Indirubin-3'-monoxime | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-08-01 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
3P0W
 
 | | Crystal structure of D-Glucarate dehydratase from Ralstonia solanacearum complexed with Mg and D-glucarate | | Descriptor: | D-GLUCARATE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of D-Glucarate dehydratase from Ralstonia solanacearum complexed with Mg and D-glucarate
To be Published
|
|
1PRM
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1PRL
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
3P60
 
 | | Crystal structure of the mutant T159V of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
1PJR
 
 | |
3P3I
 
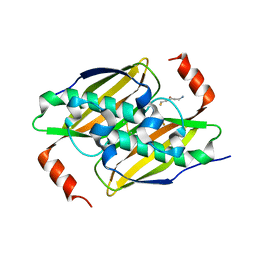 | | Crystal structure of the F36A mutant of the fluoroacetyl-CoA-specific thioesterase FlK in complex with fluoroacetate and CoA | | Descriptor: | COENZYME A, Fluoroacetyl coenzyme A thioesterase, fluoroacetic acid | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-04 | | Release date: | 2010-10-20 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
3P5Y
 
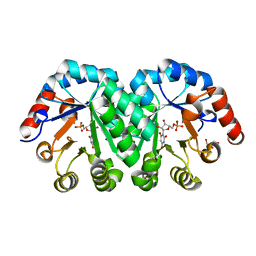 | | Crystal structure of the mutant T159A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
1QMP
 
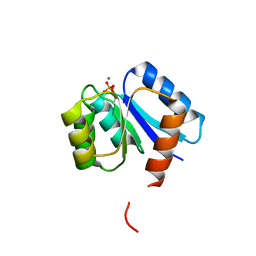 | | Phosphorylated aspartate in the crystal structure of the sporulation response regulator, Spo0A | | Descriptor: | CALCIUM ION, Stage 0 sporulation protein A | | Authors: | Lewis, R.J, Brannigan, J.A, Muchova, K, Barak, I, Wilkinson, A.J. | | Deposit date: | 1999-10-04 | | Release date: | 1999-11-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylated aspartate in the structure of a response regulator protein.
J. Mol. Biol., 294, 1999
|
|
1QDD
 
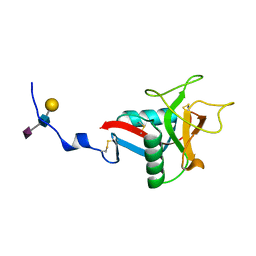 | | CRYSTAL STRUCTURE OF HUMAN LITHOSTATHINE TO 1.3 A RESOLUTION | | Descriptor: | LITHOSTATHINE, beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Gerbaud, V, Pignol, D, Loret, E, Bertrand, J.A, Berland, Y, Fontecilla-Camps, J.C, Canselier, J.P, Gabas, N, Verdier, J.M. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of calcite crystal growth inhibition by the N-terminal undecapeptide of lithostathine.
J.Biol.Chem., 275, 2000
|
|
1QGY
 
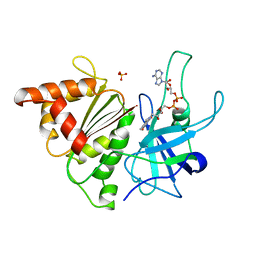 | | Ferredoxin:NADP+ reductase mutant with Lys 75 replaced by Glu (K75E) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP+ reductase, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of interactions for complex formation between Ferredoxin-NADP+ reductase and its protein partners
Proteins, 59, 2005
|
|
3RR1
 
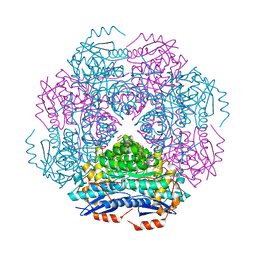 | | Crystal structure of enolase PRK14017 (target EFI-500653) from Ralstonia pickettii 12J | | Descriptor: | CHLORIDE ION, D-MALATE, Putative D-galactonate dehydratase | | Authors: | Patskovsky, Y, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of enolase PRK14017 from Ralstonia pickettii
To be Published
|
|
1QO5
 
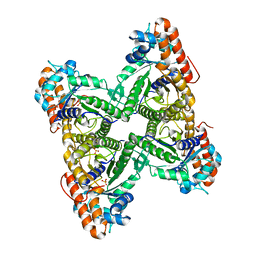 | |
1QVL
 
 | | Structure of the antimicrobial hexapeptide cyc-(RRWWRF) bound to SDS micelles | | Descriptor: | c-RW | | Authors: | Appelt, C, Soderhall, J.A, Bienert, M, Dathe, M, Schmieder, P. | | Deposit date: | 2003-08-28 | | Release date: | 2004-09-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the Antimicrobial, Cationic Hexapeptide Cyclo(RRWWRF) and Its Analogues in Solution and Bound to Detergent Micelles
Chembiochem, 6, 2005
|
|
1SO4
 
 | | Crystal structure of K64A mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-06-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
1SJ4
 
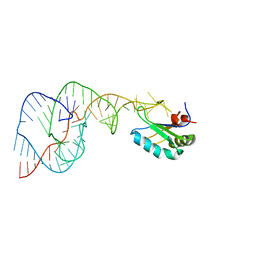 | | Crystal structure of a C75U mutant Hepatitis Delta Virus ribozyme precursor, in Cu2+ solution | | Descriptor: | precursor form of the Hepatitis Delta virus ribozyme, small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H, Doudna, J.A. | | Deposit date: | 2004-03-02 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme
catalysis
Nature, 429, 2004
|
|
1SJB
 
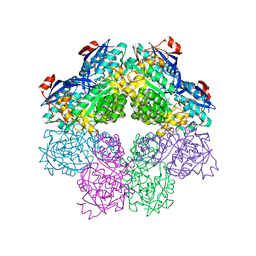 | | X-ray structure of o-succinylbenzoate synthase complexed with o-succinylbenzoic acid | | Descriptor: | 2-SUCCINYLBENZOATE, MAGNESIUM ION, N-acylamino acid racemase | | Authors: | Thoden, J.B, Taylor-Ringia, E.A, Garrett, J.B, Gerlt, J.A, Holden, H.M, Rayment, I. | | Deposit date: | 2004-03-03 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural Studies of the Promiscuous o-Succinylbenzoate Synthase from Amycolatopsis
Biochemistry, 43, 2004
|
|
1SKL
 
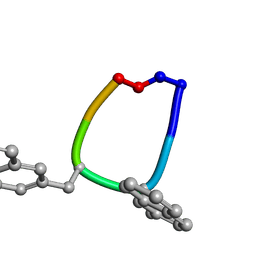 | | Structure of the antimicrobial hexapeptide cyc-(RRNalNalRF) bound to DPC micelles | | Descriptor: | cyclic hexapeptide RR(NAL)(NAL)RF | | Authors: | Appelt, C, Soderhall, J.A, Bienert, M, Dathe, M, Schmieder, P. | | Deposit date: | 2004-03-05 | | Release date: | 2005-03-15 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Structure of the antimicrobial, cationic hexapeptide cyclo(RRWWRF) and its analogues in solution and bound to detergent micelles.
Chembiochem, 6, 2005
|
|
3RR3
 
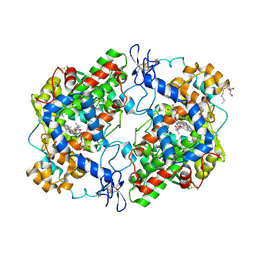 | | Structure of (R)-flurbiprofen bound to mCOX-2 | | Descriptor: | (2R)-2-(3-fluoro-4-phenyl-phenyl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Duggan, K.C, Hermanson, D.J, Musee, J, Prusakiewicz, J.J, Scheib, J, Carter, B.D, Banerjee, S, Oates, J.A, Marnett, L.J. | | Deposit date: | 2011-04-28 | | Release date: | 2011-11-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.842 Å) | | Cite: | (R)-Profens are substrate-selective inhibitors of endocannabinoid oxygenation by COX-2.
Nat.Chem.Biol., 7, 2011
|
|
3RUV
 
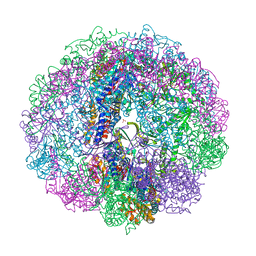 | | Crystal structure of Cpn-rls in complex with ATP analogue from Methanococcus maripaludis | | Descriptor: | Chaperonin, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
1SPD
 
 | |
