4BXE
 
 | |
1QH0
 
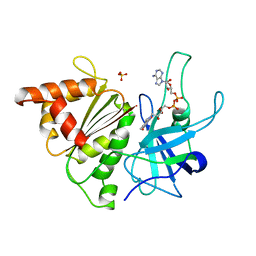 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 76 MUTATED BY ASP AND LEU 78 MUTATED BY ASP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal complex formation and electron transfer to ferredoxin.
J.Biol.Chem., 276, 2001
|
|
4BQA
 
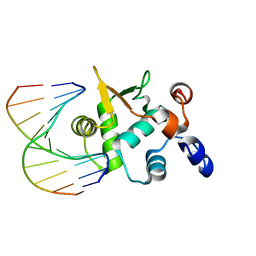 | | Crystal structure of the ETS domain of human ETS2 in complex with DNA | | Descriptor: | 5'-D(*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP)-3', 5'-D(*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP)-3', PROTEIN C-ETS-2 | | Authors: | Newman, J.A, Cooper, C.D.O, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Gileadi, O. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into the Autoregulation and Cooperativity of the Human Transcription Factor Ets-2.
J.Biol.Chem., 290, 2015
|
|
4BPR
 
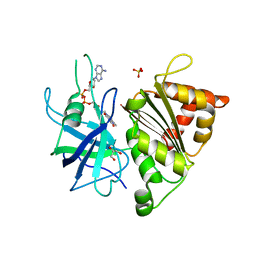 | | FERREDOXIN-NADP REDUCTASE MUTANT WITH TYR 79 REPLACED BY PHE (Y79F) | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Sanchez-Azqueta, A, Hervas, M, Navarro, J.A, Medina, M. | | Deposit date: | 2013-05-28 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Hydrogen Bond Network in the Active Site of Anabaena Ferredoxin-Nadp(+) Reductase Modulates its Catalytic Efficiency.
Biochim.Biophys.Acta, 1837, 2013
|
|
1QM4
 
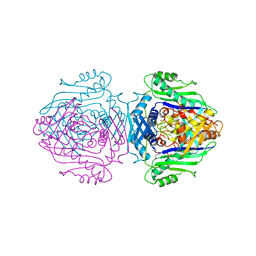 | | Methionine Adenosyltransferase Complexed with a L-Methionine Analogue | | Descriptor: | L-2-AMINO-4-METHOXY-CIS-BUT-3-ENOIC ACID, MAGNESIUM ION, METHIONINE ADENOSYLTRANSFERASE, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 1999-09-20 | | Release date: | 2000-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The Crystal Structure of Tetrameric Methionine Adenosyltransferase from Rat Liver Reveals the Methionine-Binding Site
J.Mol.Biol., 300, 2000
|
|
3OX3
 
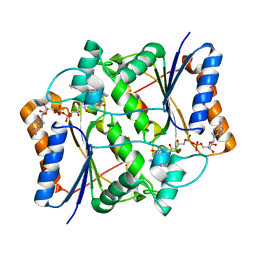 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(2-methoxy-1H-dipyrido[2,3-a:3',2'-e]pyrrolizin-11-yl)ethyl]furan-2-carboxamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
1QTW
 
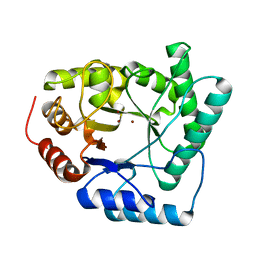 | | HIGH-RESOLUTION CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI DNA REPAIR ENZYME ENDONUCLEASE IV | | Descriptor: | ENDONUCLEASE IV, ZINC ION | | Authors: | Hosfield, D.J, Guan, Y, Haas, B.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1999-06-29 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis.
Cell(Cambridge,Mass.), 98, 1999
|
|
1MDU
 
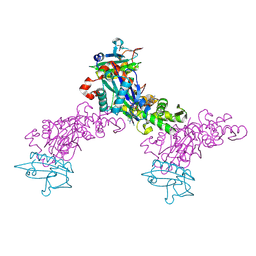 | | Crystal structure of the chicken actin trimer complexed with human gelsolin segment 1 (GS-1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Dawson, J.F, Sablin, E.P, Spudich, J.A, Fletterick, R.J. | | Deposit date: | 2002-08-07 | | Release date: | 2003-01-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an F-actin trimer disrupted by gelsolin and implications for the mechanism of severing
J.Biol.Chem., 278, 2003
|
|
3ORX
 
 | | PDK1 mutant bound to allosteric disulfide fragment inhibitor 1F8 | | Descriptor: | 2-methyl-N-(2-sulfanylethyl)-1-benzofuran-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1, CHLORIDE ION | | Authors: | Sadowsky, J.D, Wells, J.A. | | Deposit date: | 2010-09-08 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2044 Å) | | Cite: | Turning a protein kinase on or off from a single allosteric site via disulfide trapping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3P22
 
 | | Crystal structure of the ENE, a viral RNA stability element, in complex with A9 RNA | | Descriptor: | Core ENE hairpin from KSHV PAN RNA, oligo(A)9 RNA | | Authors: | Mitton-Fry, R.M, DeGregorio, S.J, Wang, J, Steitz, T.A, Steitz, J.A. | | Deposit date: | 2010-10-01 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Poly(A) tail recognition by a viral RNA element through assembly of a triple helix.
Science, 330, 2010
|
|
4J9X
 
 | | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with trans-4-hydroxy-l-proline | | Descriptor: | 4-HYDROXYPROLINE, Proline racemase family protein, SODIUM ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with trans-4-hydroxy-l-proline
To be Published
|
|
1BR8
 
 | | IMPLICATIONS FOR FUNCTION AND THERAPY OF A 2.9A STRUCTURE OF BINARY-COMPLEXED ANTITHROMBIN | | Descriptor: | PROTEIN (ANTITHROMBIN-III), PROTEIN (PEPTIDE) | | Authors: | Skinner, R, Chang, W.S.W, Jin, L, Pei, X.Y, Huntington, J.A, Abrahams, J.P, Carrell, R.W, Lomas, D.A. | | Deposit date: | 1998-08-26 | | Release date: | 1998-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Implications for function and therapy of a 2.9 A structure of binary-complexed antithrombin.
J.Mol.Biol., 283, 1998
|
|
1BN6
 
 | | HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1998-07-31 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
4JDP
 
 | | Crystal structure of probable p-nitrophenyl phosphatase (pho2) (target EFI-501307) from Archaeoglobus fulgidus DSM 4304 with Magnesium bound | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, p-nitrophenyl phosphatase (Pho2) | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Allen, K.N, Dunaway-Mariano, D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of probable p-nitrophenyl phosphatase from Archaeoglobus fulgidus.
To be Published
|
|
1QET
 
 | | TANDEM GU MISMATCHES IN RNA, NMR, 30 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*AP*UP*GP*UP*CP*C)-3') | | Authors: | Mcdowell, J.A, He, L, Chen, X, Turner, D.H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2.
Biochemistry, 36, 1997
|
|
4AF0
 
 | | Crystal structure of cryptococcal inosine monophosphate dehydrogenase | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID, MYCOPHENOLIC ACID, ... | | Authors: | Valkov, E, Stamp, A, Morrow, C.A, Kobe, B, Fraser, J.A. | | Deposit date: | 2012-01-15 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De Novo GTP Biosynthesis is Critical for Virulence of the Fungal Pathogen Cryptococcus Neoformans
Plos Pathog., 8, 2012
|
|
3OS8
 
 | | Estrogen Receptor | | Descriptor: | 4-[1-benzyl-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor | | Authors: | Bruning, J, Parent, A.A, Gil, G, Zhao, M, Nowak, J, Pace, M.C, Smith, C.L, Afonine, P.V, Adams, P.D, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-10 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Coupling of receptor conformation and ligand orientation determine graded activity.
Nat.Chem.Biol., 6, 2010
|
|
4JFX
 
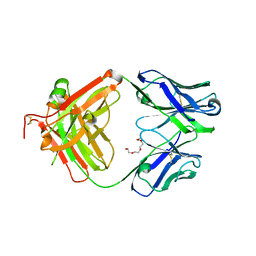 | | Structure of phosphotyrosine (pTyr) scaffold bound to pTyr peptide | | Descriptor: | Fab heavy chain, Fab light chain, Phosphopeptide, ... | | Authors: | Koerber, J.T, Thomsen, N.D, Hannigan, B.T, Degrado, W.F, Wells, J.A. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Nature-inspired design of motif-specific antibody scaffolds.
Nat.Biotechnol., 31, 2013
|
|
4JQZ
 
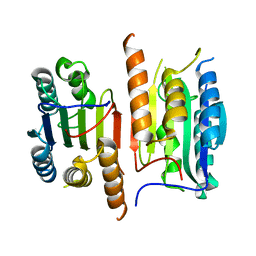 | | Human procaspase-3, crystal form 2 | | Descriptor: | Procaspase-3 | | Authors: | Thomsen, N.D, Wells, J.A. | | Deposit date: | 2013-03-20 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | Structural snapshots reveal distinct mechanisms of procaspase-3 and -7 activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3OVM
 
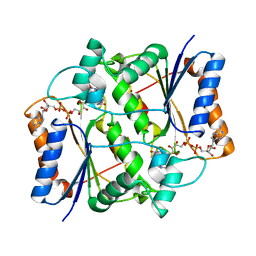 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION, ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A. | | Deposit date: | 2010-09-16 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
3P3B
 
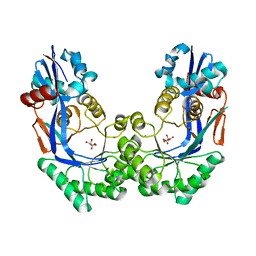 | | CRYSTAL STRUCTURE OF Galacturonate DEHYDRATASE FROM GEOBACILLUS SP. COMPLEXED WITH D-TARTRATE | | Descriptor: | D(-)-TARTARIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Groninger-Poe, F, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-04 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | CRYSTAL STRUCTURE OF Galacturonate DEHYDRATASE FROM GEOBACILLUS SP. COMPLEXED WITH D-TARTRATE.
To be Published
|
|
3P61
 
 | | Crystal structure of the mutant R160A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
1BN7
 
 | | HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | ACETATE ION, HALOALKANE DEHALOGENASE | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1998-07-31 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
4A8N
 
 | | Protein crystallization and microgravity: glucose isomerase crystals grown during the PCDF-PROTEIN mission | | Descriptor: | COBALT (II) ION, GLYCEROL, XYLOSE ISOMERASE | | Authors: | Decanniere, K, Patino-Lopez, L.-D, Sleutel, M, Evrard, C, Van De Weerdt, C, Haumont, E, Gavira, J.A, Otalora, F, Maes, D. | | Deposit date: | 2011-11-21 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protein Crystallization and Microgravity: Glucose Isomerase Crystals Grown During the Pcdf-Protein Mission
To be Published
|
|
1MUD
 
 | | CATALYTIC DOMAIN OF MUTY FROM ESCHERICHIA COLI, D138N MUTANT COMPLEXED TO ADENINE | | Descriptor: | ADENINE, Adenine DNA glycosylase, GLYCEROL, ... | | Authors: | Guan, Y, Tainer, J.A. | | Deposit date: | 1998-08-20 | | Release date: | 1999-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily.
Nat.Struct.Biol., 5, 1998
|
|
