5UCE
 
 | | Solution NMR structure of the major species of DANCER-2, a dynamic and natively folded pentamutant of the B1 domain of streptococcal protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Damry, A.M, Davey, J.A, Goto, N.K, Chica, R.A. | | Deposit date: | 2016-12-22 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational design of proteins that exchange on functional timescales.
Nat. Chem. Biol., 13, 2017
|
|
5V5B
 
 | | KVQIINKKLD, Structure of the amyloid spine from microtubule associated protein tau Repeat 2 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Seidler, P.M, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S, Cascio, D, Boyer, D.R. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.5 Å) | | Cite: | Structure-based inhibitors of tau aggregation.
Nat Chem, 10, 2018
|
|
5UP9
 
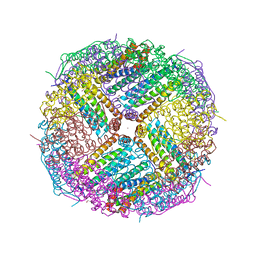 | | Crystal Structure of Zn-bound Human Heavy-Chain ferritin variant 122H-delta C-star with para-xylenedihydroxamate | | Descriptor: | 2,2'-(1,4-phenylene)bis(N-hydroxyacetamide), DI(HYDROXYETHYL)ETHER, Ferritin heavy chain, ... | | Authors: | Bailey, J.B, Zhang, L, Chiong, J.A, Ahn, S, Tezcan, F.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthetic Modularity of Protein-Metal-Organic Frameworks.
J. Am. Chem. Soc., 139, 2017
|
|
5V4L
 
 | |
5V5C
 
 | | VQIINK, Structure of the amyloid-spine from microtubule associated protein tau Repeat 2 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Seidler, P.M, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S, Cascio, D, Boyer, D.R. | | Deposit date: | 2017-03-14 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.25 Å) | | Cite: | Structure-based inhibitors of tau aggregation.
Nat Chem, 10, 2018
|
|
3IVK
 
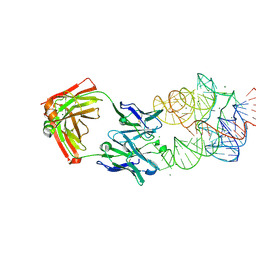 | | Crystal Structure of the Catalytic Core of an RNA Polymerase Ribozyme Complexed with an Antigen Binding Antibody Fragment | | Descriptor: | CADMIUM ION, CHLORIDE ION, Fab heavy chain, ... | | Authors: | Koldobskaya, Y, Duguid, E.M, Shechner, D.M, Koide, S, Kossiakoff, A.A, Bartel, D.P, Piccirilli, J.A. | | Deposit date: | 2009-09-01 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
5UI9
 
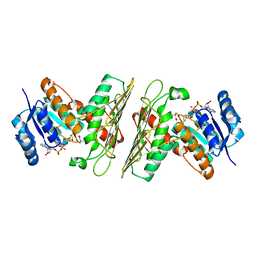 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, 2 -hydroxy-2-hydroxymethyl propanoic acid and Magnesium | | Descriptor: | (2S)-2,3-dihydroxy-2-methylpropanoic acid, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, 2 -hydroxy-2-hydroxymethyl propanoic acid and Magnesium
To be published
|
|
1SDE
 
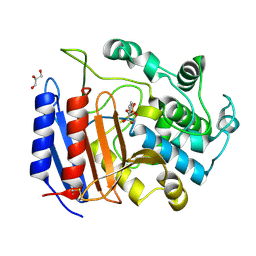 | | Toward Better Antibiotics: Crystal Structure Of D-Ala-D-Ala Peptidase inhibited by a novel bicyclic phosphate inhibitor | | Descriptor: | 2-[(DIOXIDOPHOSPHINO)OXY]BENZOATE, D-alanyl-D-alanine carboxypeptidase, GLYCEROL | | Authors: | Silvaggi, N.R, Kaur, K, Adediran, S.A, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2004-02-13 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Toward better antibiotics: crystallographic studies of a novel class of DD-peptidase/beta-lactamase inhibitors
Biochemistry, 43, 2004
|
|
3IZG
 
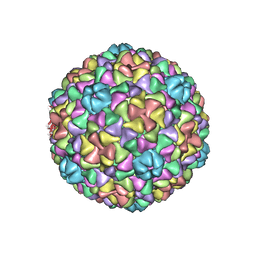 | | Bacteriophage T7 prohead shell EM-derived atomic model | | Descriptor: | Major capsid protein 10A | | Authors: | Ionel, A, Velazquez-Muriel, J.A, Agirrezabala, X, Luque, D, Cuervo, A, Caston, J.R, Valpuesta, J.M, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2010-10-27 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7.
J.Biol.Chem., 286, 2011
|
|
5V33
 
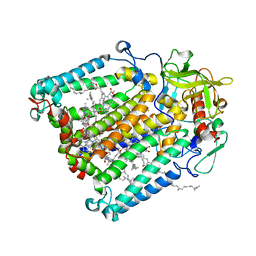 | | R. sphaeroides photosythetic reaction center mutant - Residue L223, Ser to Trp - Room Temperature Structure Solved on X-ray Transparent Microfluidic Chip | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Schieferstein, J.M, Pawate, A.S, Sun, C, Wan, F, Broecker, J, Ernst, O.P, Gennis, R.B, Kenis, P.J.A. | | Deposit date: | 2017-03-06 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.487 Å) | | Cite: | X-ray transparent microfluidic chips for high-throughput screening and optimization of in meso membrane protein crystallization.
Biomicrofluidics, 11, 2017
|
|
5UK9
 
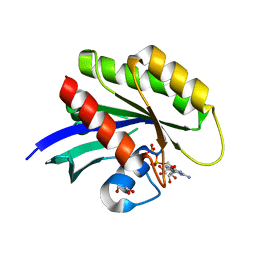 | | Wild-type K-Ras(GCP) pH 6.5 | | Descriptor: | GLYCEROL, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Parker, J.A, Mattos, C. | | Deposit date: | 2017-01-20 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | K-Ras Populates Conformational States Differently from Its Isoform H-Ras and Oncogenic Mutant K-RasG12D.
Structure, 26, 2018
|
|
1PWG
 
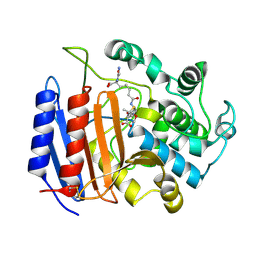 | | Covalent Penicilloyl Acyl Enzyme Complex Of The Streptomyces R61 DD-Peptidase With A Highly Specific Penicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(6S)-6-carboxy-6-(glycylamino)hexanoyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.074 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
5DR9
 
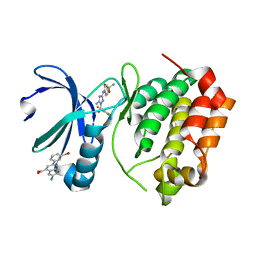 | | Aurora A Kinase in Complex with AA29 and JNJ-7706621 in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)-6-chloroquinoline-4-carboxylic acid, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Aurora kinase A | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5DPV
 
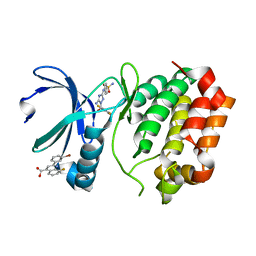 | | Aurora A Kinase in Complex with AA35 and JNJ-7706621 in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)-8-fluoroquinoline-4-carboxylic acid, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Aurora kinase A | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5DT0
 
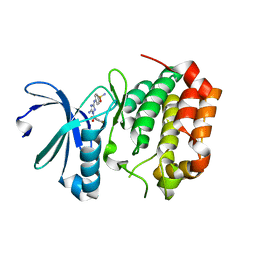 | | Aurora A Kinase in Complex with JNJ-7706621 in Space Group P6122 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Aurora kinase A | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
2KUB
 
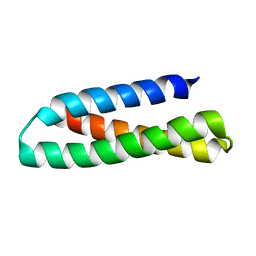 | | Solution structure of the alpha subdomain of the major non-repeat unit of Fap1 fimbriae of Streptococcus parasanguis | | Descriptor: | Fimbriae-associated protein Fap1 | | Authors: | Ramboarina, S, Garnett, J.A, Bodey, A, Simpson, P, Bardiaux, B, Nilges, M, Matthews, S. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into serine-rich fimbriae from gram-positive bacteria.
J.Biol.Chem., 2010
|
|
5DN3
 
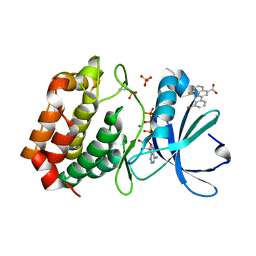 | | Aurora A in complex with ATP and AA35. | | Descriptor: | 2-(3-bromophenyl)-8-fluoroquinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G, Huggins, D, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A. | | Deposit date: | 2015-09-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5DR2
 
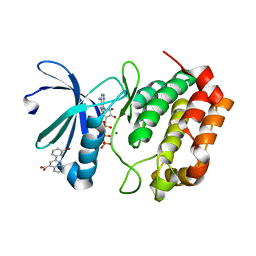 | | Aurora A Kinase in Complex with AA30 and ATP in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)quinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5DT4
 
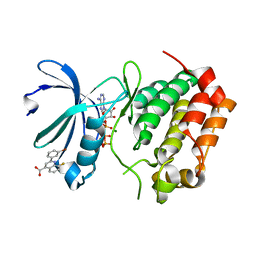 | | Aurora A Kinase in Complex with AA35 and ATP in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)-8-fluoroquinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
2L67
 
 | | Solution Structure of Human Apo L-FABP | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
6TAQ
 
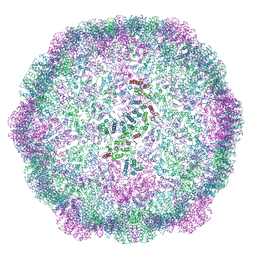 | |
6TAU
 
 | |
6TAR
 
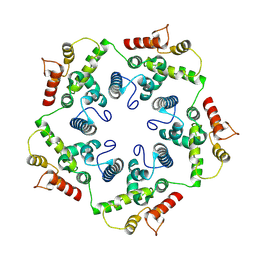 | |
8RB4
 
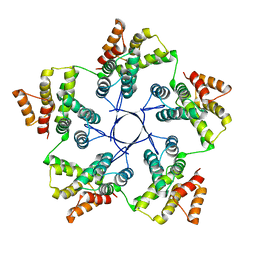 | |
6T64
 
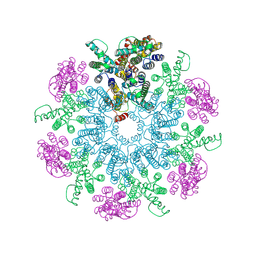 | | A model of the EIAV CA-SP hexamer (C6) from Gag-deltaMA spheres assembled at pH6 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
