5SDH
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z2856434854 | | Descriptor: | 1-[(3-methoxyphenyl)methyl]piperidine-4-carboxamide, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDC
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z2856434912 | | Descriptor: | 3-[(4-methylpiperidin-1-yl)methyl]-1H-indole, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDO
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z19735067 | | Descriptor: | 2-(4-fluorophenoxy)-1-(pyrrolidin-1-yl)ethan-1-one, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDL
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z321318226 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, N-(4-methoxyphenyl)-N'-pyridin-4-ylurea | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDR
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z1273312153 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, N-methyl-1H-indole-7-carboxamide | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDI
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z44592329 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, N-phenyl-N'-pyridin-3-ylurea | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDP
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z2277255954 | | Descriptor: | 5-fluoro-1-(1-methylcyclopropane-1-sulfonyl)-1,2,3,6-tetrahydropyridine, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDF
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z2856434834 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, N-(2,3-dimethylphenyl)-2-(morpholin-4-yl)acetamide | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDN
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z437584380 | | Descriptor: | (4-chlorophenyl)(thiomorpholin-4-yl)methanone, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5TW8
 
 | |
1IKI
 
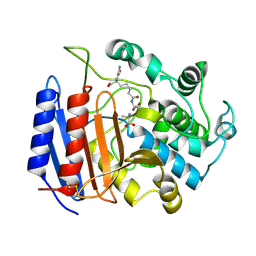 | | COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH THE PRODUCTS OF A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANINE, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
5TYF
 
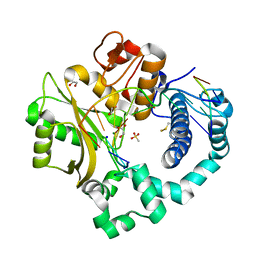 | | DNA Polymerase Mu Product Complex, 10 mM Mg2+ (270 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-19 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
5TYW
 
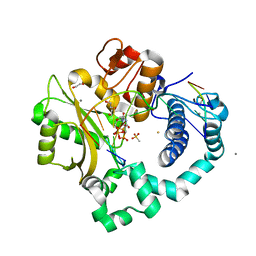 | | DNA Polymerase Mu Reactant Complex, Mn2+ (10 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
5TOD
 
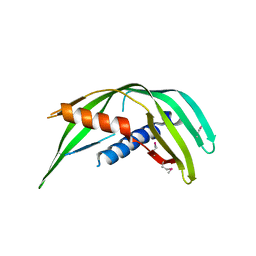 | | Transmembrane protein 24 SMP domain | | Descriptor: | Transmembrane protein 24 | | Authors: | Lees, J.A, Reinisch, K.M. | | Deposit date: | 2016-10-17 | | Release date: | 2017-03-01 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion.
Science, 355, 2017
|
|
5TP9
 
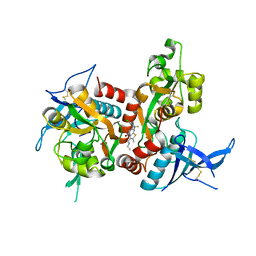 | | Structure of the human GluN1/GluN2A LBD in complex with compound 2 (GNE9178) | | Descriptor: | 7-{[5-chloro-3-(trifluoromethyl)-1H-pyrazol-1-yl]methyl}-N-ethyl-2-methyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | GluN2A-Selective Pyridopyrimidinone Series of NMDAR Positive Allosteric Modulators with an Improved in Vivo Profile.
ACS Med Chem Lett, 8, 2017
|
|
2OQC
 
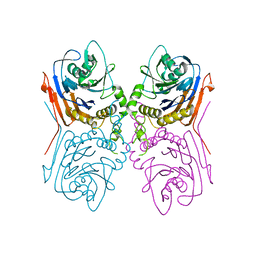 | | Crystal Structure of Penicillin V acylase from Bacillus subtilis | | Descriptor: | Penicillin V acylase | | Authors: | Suresh, C.G, Rathinaswamy, P, Pundle, A.V, Prabhune, A.A, Sivaraman, H, Brannigan, J.A, Dodson, G.G. | | Deposit date: | 2007-01-31 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Penicillin V acylase from Bacillus subtilis
to be published
|
|
2OLP
 
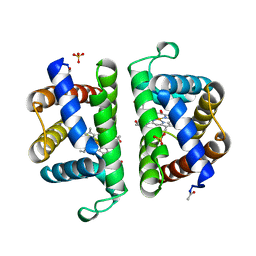 | | Structure and ligand selection of hemoglobin II from Lucina pectinata | | Descriptor: | Hemoglobin II, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gavira, J.A, Camara-Artigas, A, de Jesus, W, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | Deposit date: | 2007-01-19 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structure and Ligand Selection of Hemoglobin II from Lucina pectinata
J.Biol.Chem., 283, 2008
|
|
1N3B
 
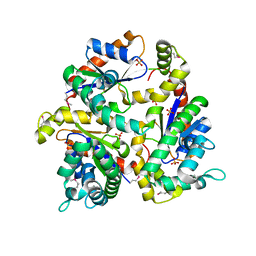 | | Crystal Structure of Dephosphocoenzyme A kinase from Escherichia coli | | Descriptor: | Dephospho-CoA kinase, SULFATE ION | | Authors: | O'Toole, N, Barbosa, J.A.R.G, Li, Y, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-28 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Trimeric Form of Dephosphocoenzyme A Kinase from Escherichia coli
Protein Sci., 12, 2003
|
|
5H8H
 
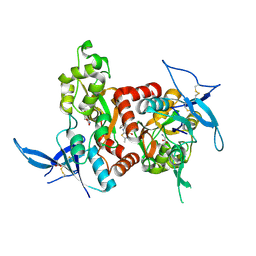 | | Structure of the human GluN1/GluN2A LBD in complex with GNE3419 | | Descriptor: | 7-[[ethyl(phenyl)amino]methyl]-2-methyl-[1,3,4]thiadiazolo[3,2-a]pyrimidin-5-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
5TYB
 
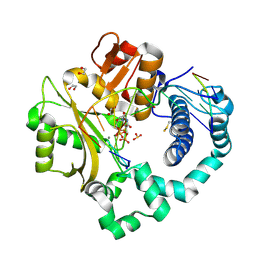 | | DNA Polymerase Mu Reactant Complex, 10mM Mg2+ (7.5 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-19 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
5TY2
 
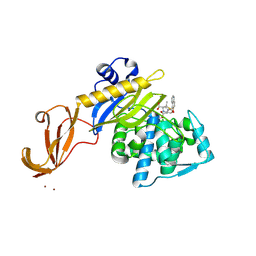 | | Crystal structure of S. aureus penicillin binding protein 4 (PBP4) mutant (E183A, F241R) in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-18 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
5TYI
 
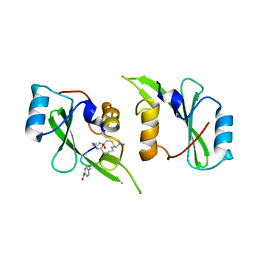 | | Grb7 SH2 with bicyclic peptide containing pY mimetic | | Descriptor: | Growth factor receptor-bound protein 7, Peptide inhibitor | | Authors: | Watson, G.M, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2016-11-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
2SSP
 
 | | LEUCINE-272-ALANINE URACIL-DNA GLYCOSYLASE BOUND TO ABASIC SITE-CONTAINING DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*(AAB)P*AP*TP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
5TYV
 
 | | DNA Polymerase Mu Reactant Complex, Mn2+ (7.5 min) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
5DTW
 
 | |
