1SJC
 
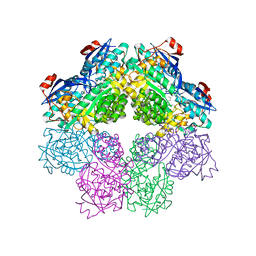 | | x-ray structure of o-succinylbenzoate synthase complexed with N-succinyl methionine | | Descriptor: | MAGNESIUM ION, N-SUCCINYL METHIONINE, N-acylamino acid racemase | | Authors: | Thoden, J.B, Taylor-Ringia, E.A, Garrett, J.B, Gerlt, J.A, Holden, H.M, Rayment, I. | | Deposit date: | 2004-03-03 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural Studies of the Promiscuous o-Succinylbenzoate Synthase from Amycolatopsis
Biochemistry, 43, 2004
|
|
1SKI
 
 | | Structure of the antimicrobial hexapeptide cyc-(RRYYRF) bound to DPC micelles | | Descriptor: | cyclic hexapeptide RRYYRF | | Authors: | Appelt, C, Soderhall, J.A, Bienert, M, Dathe, M, Schmieder, P. | | Deposit date: | 2004-03-05 | | Release date: | 2005-03-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the antimicrobial, cationic hexapeptide cyclo(RRWWRF) and its analogues in solution and bound to detergent micelles.
Chembiochem, 6, 2005
|
|
3QKS
 
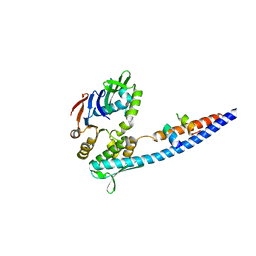 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1ET9
 
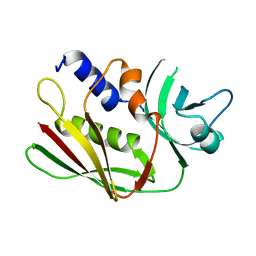 | | CRYSTAL STRUCTURE OF THE SUPERANTIGEN SPE-H FROM STREPTOCOCCUS PYOGENES | | Descriptor: | SUPERANTIGEN SPE-H | | Authors: | Arcus, V.L, Proft, T, Sigrell, J.A, Baker, H.M, Fraser, J.D, Baker, E.N. | | Deposit date: | 2000-04-12 | | Release date: | 2000-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes.
J.Mol.Biol., 299, 2000
|
|
4B1J
 
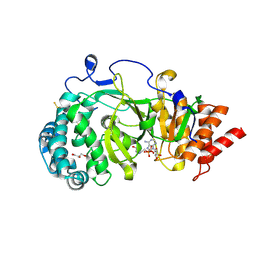 | | Structure of human PARG catalytic domain in complex with ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, GLYCEROL, POLY(ADP-RIBOSE) GLYCOHYDROLASE, ... | | Authors: | Brassington, C, Ellston, J, Hassall, G, Holdgate, G, McAlister, M, Overman, R, Smith, G, Tucker, J.A, Watson, M. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of the Human Poly (Adp-Ribose) Glycohydrolase Catalytic Domain Confirm Catalytic Mechanism and Explain Inhibition by Adp-Hpd Derivatives.
Plos One, 7, 2012
|
|
4B88
 
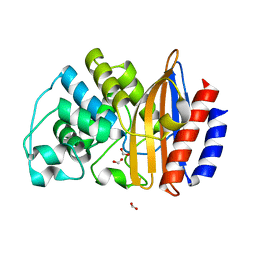 | |
4GH9
 
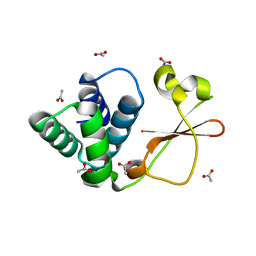 | | Crystal structure of Marburg virus VP35 RNA binding domain | | Descriptor: | ACETATE ION, Polymerase cofactor VP35 | | Authors: | Bale, S, Jean-Philippe, J, Bornholdt, Z.A, Kimberlin, C.K, Halfmann, P, Zandonatti, M.A, Kunert, J, Kroon, G.J.A, Kawaoka, Y, MacRae, I.J, Wilson, I.A, Saphire, E.O. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Marburg Virus VP35 Can Both Fully Coat the Backbone and Cap the Ends of dsRNA for Interferon Antagonism.
Plos Pathog., 8, 2012
|
|
4B1H
 
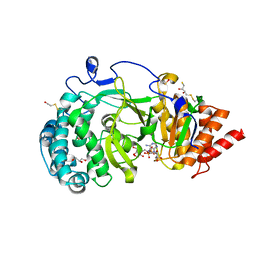 | | Structure of human PARG catalytic domain in complex with ADP-ribose | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Brassington, C, Ellston, J, Hassall, G, Holdgate, G, McAlister, M, Smith, G, Tucker, J.A, Watson, M. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Human Poly (Adp-Ribose) Glycohydrolase Catalytic Domain Confirm Catalytic Mechanism and Explain Inhibition by Adp-Hpd Derivatives.
Plos One, 7, 2012
|
|
4LE8
 
 | |
1E64
 
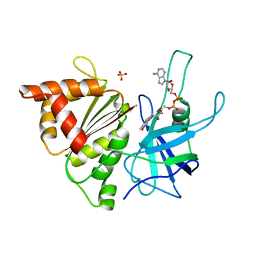 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LYS 75 REPLACED BY GLN (K75Q) | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2000-08-07 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners.
Proteins, 59, 2005
|
|
1R6W
 
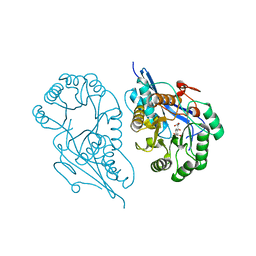 | | Crystal structure of the K133R mutant of o-Succinylbenzoate synthase (OSBS) from Escherichia coli. Complex with SHCHC | | Descriptor: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, MAGNESIUM ION, o-Succinylbenzoate Synthase | | Authors: | Klenchin, V.A, Taylor Ringia, E.A, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-10-17 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural and Mutagenic Studies of the Mechanism of the Reaction Catalyzed by o-Succinylbenzoate Synthase from Escherichia coli
Biochemistry, 42, 2003
|
|
4B1I
 
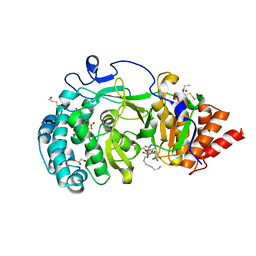 | | Structure of human PARG catalytic domain in complex with OA-ADP-HPD | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 8-n-octylamino-adenosine diphosphate hydroxypyrrolidinediol, GLYCEROL, ... | | Authors: | Brassington, C, Ellston, J, Hassall, G, Holdgate, G, Johnson, T, McAlister, M, Smith, G, Tucker, J.A, Watson, M. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of the Human Poly (Adp-Ribose) Glycohydrolase Catalytic Domain Confirm Catalytic Mechanism and Explain Inhibition by Adp-Hpd Derivatives.
Plos One, 7, 2012
|
|
4BEQ
 
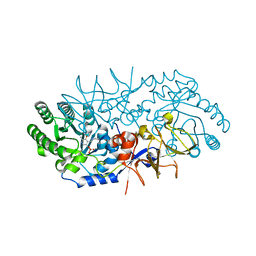 | |
1M1K
 
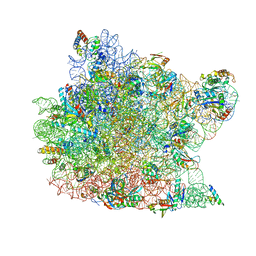 | | Co-crystal structure of azithromycin bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, AZITHROMYCIN, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2002-06-19 | | Release date: | 2002-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
4B8R
 
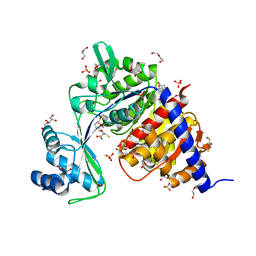 | | Crystal Structure of Thermococcus litoralis ADP-dependent Glucokinase (GK) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADP-DEPENDENT GLUCOKINASE, ... | | Authors: | Herrera-Morande, A, Rivas-Pardo, J.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure, Saxs and Kinetic Mechanism of Hyperthermophilic Adp-Dependent Glucokinase from Thermococcus Litoralis Reveal a Conserved Mechanism for Catalysis.
Plos One, 8, 2013
|
|
4KWD
 
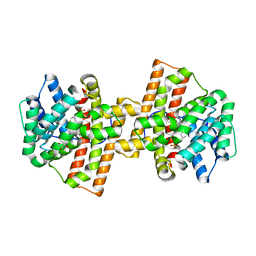 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with (1R,8R,9aS)-1,9a-dimethyl-8-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | Descriptor: | (1R,5R,8R,9aS)-1,9a-dimethyl-8-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Al-lami, N, Janvier, M, D'Antonio, E.L, Faraldos, J.A, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
4KZ2
 
 | | Crystal Structure of phi29 pRNA 3WJ Core | | Descriptor: | MANGANESE (II) ION, phi29 pRNA 3WJ core RNA 16 mer, phi29 pRNA 3WJ core RNA 18 mer, ... | | Authors: | Zhang, H, Endrizzi, J.A, Shu, Y, Haque, F, Sauter, C, Guo, P, Chi, Y.-I. | | Deposit date: | 2013-05-29 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA.
Rna, 19, 2013
|
|
1ME3
 
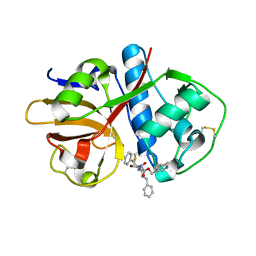 | |
4BJ4
 
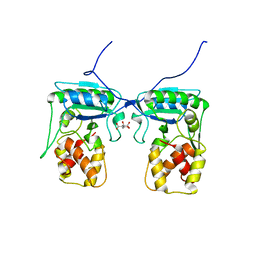 | | Structure of Pseudomonas aeruginosa amidase Ampdh2 | | Descriptor: | AMPDH2, CITRATE ANION | | Authors: | Martinez-Caballero, C.S, Carrasco-Lopez, C, Artola-Recolons, C, Hermoso, J.A. | | Deposit date: | 2013-04-16 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
1R9G
 
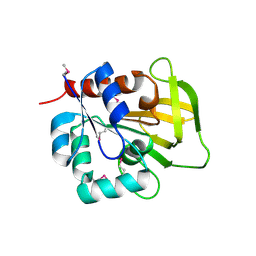 | | Three-dimensional Structure of YaaE from Bacillus subtilis | | Descriptor: | Hypothetical protein yaaE | | Authors: | Bauer, J.A, Bennett, E.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2003-10-29 | | Release date: | 2004-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional Structure of YaaE from Bacillus subtilis, a Glutaminase Implicated in Pyridoxal-5'-phosphate Biosynthesis.
J.Biol.Chem., 279, 2004
|
|
1E82
 
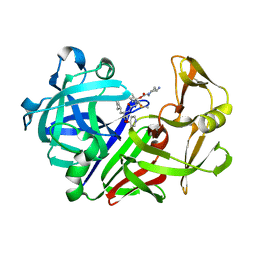 | | Endothiapepsin complex with renin inhibitor MERCK-KGAA-EMD59601 | | Descriptor: | (2S)-1-{[(2R)-1-{[(2S,3R)-1-cyclohexyl-3-hydroxy-4-(pyridin-4-yloxy)butan-2-yl]amino}-3-(methylsulfanyl)-1-oxopropan-2-yl]amino}-1-oxo-3-phenylpropan-2-yl 4-aminopiperidine-1-carboxylate, ENDOTHIAPEPSIN | | Authors: | Read, J.A, Cooper, J.B, Toldo, L, Rippmann, F, Raddatz, P. | | Deposit date: | 2000-09-15 | | Release date: | 2000-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Refinement of Four Endothiapepsin Inhibitor Complexes. Crystallographic Studies of Cytochrome Ch from Methylobacterium Extorquens and Inhibitor Complexes of Aspartic Proteinases.
Thesis, 1999
|
|
1LUW
 
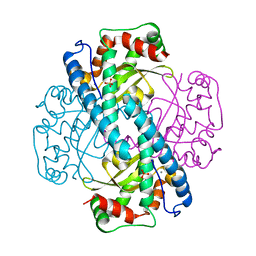 | | CATALYTIC AND STRUCTURAL EFFECTS OF AMINO-ACID SUBSTITUTION AT HIS 30 IN HUMAN MANGANESE SUPEROXIDE DISMUTASE: INSERTION OF VAL CGAMMA INTO THE SUBSTRATE ACCESS CHANNEL | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Superoxide dismutase [Mn] | | Authors: | Hearn, A.S, Stroupe, M.E, Ramilo, C.A, Luba, J.P, Cabelli, D.E, Tainer, J.A, Silverman, D.N. | | Deposit date: | 2002-05-23 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic and structural effects of amino acid substitution at histidine 30 in human manganese superoxide dismutase: insertion of valine C gamma into the substrate access channel
Biochemistry, 42, 2003
|
|
4B12
 
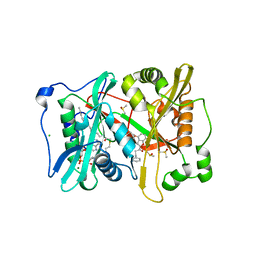 | | Plasmodium vivax N-myristoyltransferase with a bound benzofuran inhibitor (compound 23) | | Descriptor: | 1-[3-methyl-4-(piperidin-4-yloxy)-1-benzofuran-2-yl]-3-phenylpropan-1-one, 2-oxopentadecyl-CoA, CHLORIDE ION, ... | | Authors: | Yu, Z, Brannigan, J.A, Moss, D.K, Brzozowski, A.M, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Design and Synthesis of Inhibitors of Plasmodium Falciparum N-Myristoyltransferase, a Promising Target for Anti-Malarial Drug Discovery.
J.Med.Chem., 55, 2012
|
|
1ED3
 
 | | CRYSTAL STRUCTURE OF RAT MINOR HISTOCOMPATIBILITY ANTIGEN COMPLEX RT1-AA/MTF-E. | | Descriptor: | BETA-2-MICROGLOBULIN, CLASS I MAJOR HISTOCOMPATIBILITY ANTIGEN RT1-AA, PEPTIDE MTF-E (13N3E) | | Authors: | Speir, J.A, Stevens, J, Joly, E, Butcher, G.W, Wilson, I.A. | | Deposit date: | 2000-01-26 | | Release date: | 2001-02-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two different, highly exposed, bulged structures for an unusually long peptide bound to rat MHC class I RT1-Aa.
Immunity, 14, 2001
|
|
1EF9
 
 | | THE CRYSTAL STRUCTURE OF METHYLMALONYL COA DECARBOXYLASE COMPLEXED WITH 2S-CARBOXYPROPYL COA | | Descriptor: | 2-CARBOXYPROPYL-COENZYME A, METHYLMALONYL COA DECARBOXYLASE | | Authors: | Benning, M.M, Haller, T, Gerlt, J.A, Holden, H.M. | | Deposit date: | 2000-02-07 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | New reactions in the crotonase superfamily: structure of methylmalonyl CoA decarboxylase from Escherichia coli.
Biochemistry, 39, 2000
|
|
