4O8R
 
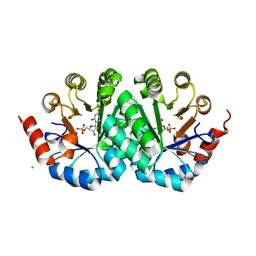 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 5,6-dihydrouridine 5'-monophosphate | | Descriptor: | 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, CHLORIDE ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-12-29 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 5,6-dihydrouridine 5'-monophosphate
To be Published
|
|
4OY5
 
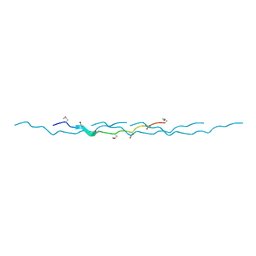 | | 0.89 Angstrom resolution crystal structure of (Gly-Pro-Hyp)10 | | Descriptor: | Collagen | | Authors: | Suzuki, H, Mahapatra, D, Steel, P.J, Dyer, J, Dobson, R.C.J, Gerrard, J.A, Valery, C. | | Deposit date: | 2014-02-10 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Sub-angstrom structure of the collagen model peptide (GPO)10 shows a hydrated triple helix with pitch variation and two proline ring conformations
To Be Published
|
|
4QF4
 
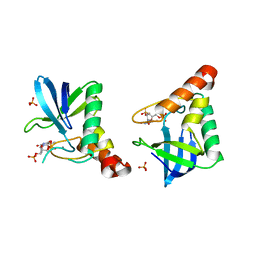 | | Crystal structure of Staphylococcal nuclease variant V23M at cryogenic temperature | | Descriptor: | CALCIUM ION, PHOSPHATE ION, THYMIDINE-3',5'-DIPHOSPHATE, ... | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno, E.B. | | Deposit date: | 2014-05-19 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4QAY
 
 | |
4Q3Z
 
 | |
2J8G
 
 | |
4Q2H
 
 | | Crystal structure of probable proline racemase from agrobacterium radiobacter K84, TARGET EFI-506561, with bound carbonate | | Descriptor: | BICARBONATE ION, GLYCEROL, Proline racemase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Proline Racemase Arad_0731 from Agrobacterium Radiobacter, Target Efi-506561
To be Published
|
|
2J8F
 
 | |
2IXU
 
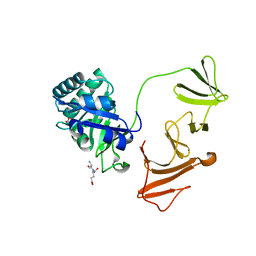 | |
8D64
 
 | | ELIC with cysteamine in POPC nanodisc | | Descriptor: | 2-AMINO-ETHANETHIOL, Erwinia ligand-gated ion channel | | Authors: | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
8D67
 
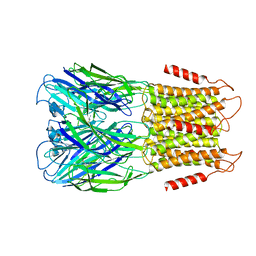 | | ELIC3 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc | | Descriptor: | 2-AMINO-ETHANETHIOL, Erwinia ligand-gated ion channel | | Authors: | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
8D66
 
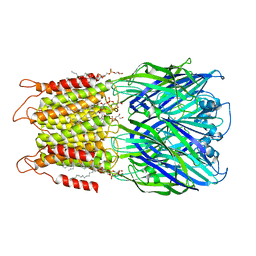 | | ELIC with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-AMINO-ETHANETHIOL, Erwinia ligand-gated ion channel | | Authors: | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
8D65
 
 | | ELIC apo in 2:1:1 POPC:POPE:POPG nanodisc | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Erwinia ligand-gated ion channel | | Authors: | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
8D63
 
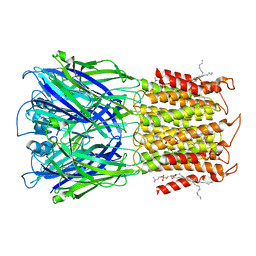 | | ELIC apo in POPC nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Erwinia ligand-gated ion channel | | Authors: | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
4RMF
 
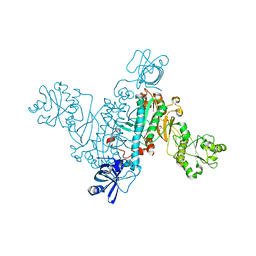 | | Biochemical and structural characterization of mycobacterial aspartyl-tRNA synthetase AspS, a promising TB drug target | | Descriptor: | 2,2-bis(hydroxymethyl)propane-1,3-diol, Aspartate--tRNA(Asp/Asn) ligase, FORMIC ACID | | Authors: | Cox, J.A.G, Gurcha, S.S, Veeraraghavan, U, Besra, G.S, Futterer, K. | | Deposit date: | 2014-10-21 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and Structural Characterization of Mycobacterial Aspartyl-tRNA Synthetase AspS, a Promising TB Drug Target.
Plos One, 9, 2014
|
|
1XEE
 
 | | Solution structure of the Chemotaxis Inhibitory Protein of Staphylococcus aureus | | Descriptor: | chemotaxis-inhibiting protein CHIPS | | Authors: | Haas, P.J, de Haas, C.J, Poppelier, M.J, van Kessel, K.P, van Strijp, J.A, Dijkstra, K, Scheek, R.M, Fan, H, Kruijtzer, J.A, Liskamp, R.M, Kemmink, J. | | Deposit date: | 2004-09-10 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the C5a receptor-blocking domain of chemotaxis inhibitory protein of Staphylococcus aureus is related to a group of immune evasive molecules
J.Mol.Biol., 353, 2005
|
|
1WAK
 
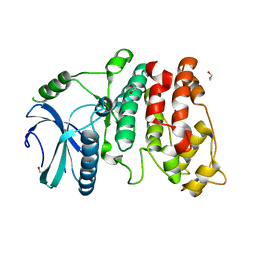 | | X-ray structure of SRPK1 | | Descriptor: | 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE SPRK1 | | Authors: | Ngo, J.C, Gullingsrud, J, Chakrabarti, S, Nolen, B, Aubol, B.E, Fu, X.D, Adams, J.A, Mccammon, J.A, Ghosh, G. | | Deposit date: | 2004-10-26 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Sr Protein Kinase 1 is Resilient to Inactivation.
Structure, 15, 2007
|
|
4TQQ
 
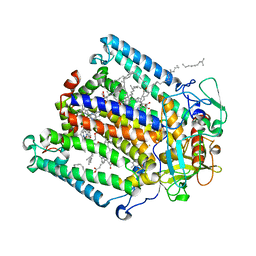 | | Photosynthetic Reaction Center from R. sphaeroides Analyzed at Room Temperature on an X-ray Transparent Microfluidic Chip | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Schieferstein, J.M, Khvostichenko, D.S, Pawate, A.S, Kenis, P.J.A. | | Deposit date: | 2014-06-11 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | X-ray Transparent Microfluidic Chip for Mesophase-Based Crystallization of Membrane Proteins and On-Chip Structure Determination.
Cryst.Growth Des., 14, 2014
|
|
2HI3
 
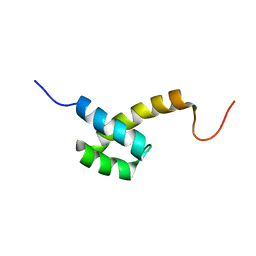 | | Solution structure of the homeodomain-only protein HOP | | Descriptor: | Homeodomain-only protein | | Authors: | Mackay, J.P, Kook, H, Epstein, J.A, Simpson, R.J, Yung, W.W. | | Deposit date: | 2006-06-28 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Analysis of the structure and function of the transcriptional coregulator HOP
Biochemistry, 45, 2006
|
|
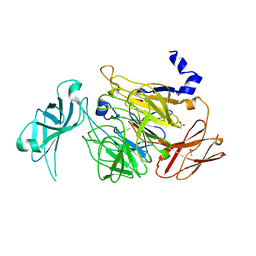
4UNM
 
 | |
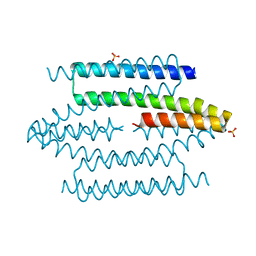
4UIG
 
 | |
4USN
 
 | | The structure of the immature HIV-1 capsid in intact virus particles at sub-nm resolution | | Descriptor: | P24 | | Authors: | Schur, F.K.M, Hagen, W.J.H, Rumlova, M, Ruml, T, Mueller, B, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structure of the Immature HIV-1 Capsid in Intact Virus Particles at 8.8 A Resolution.
Nature, 517, 2015
|
|
1YT7
 
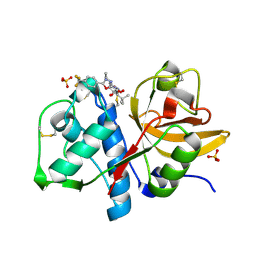 | | Cathepsin K complexed with a constrained ketoamide inhibitor | | Descriptor: | (1R)-2,2-DIMETHYL-1-({5-[4-(TRIFLUOROMETHYL)PHENYL]-1,3,4-OXADIAZOL-2-YL}METHYL)PROPYL (1S)-1-{OXO[(2-OXO-1,3-OXAZOLIDIN-3-YL)AMINO]ACETYL}PENTYLCARBAMATE, Cathepsin K, SULFATE ION | | Authors: | Barrett, D.G, Boncek, V.M, Catalano, J.G, Deaton, D.N, Hassell, A.M, Jurgensen, C.H, Long, S.T, McFadyen, R.B, Miller, A.B, Miller, L.R, Payne, J.A, Ray, J.A, Samano, V, Shewchuk, L.M, Tavares, F.X, Wells-Knecht, K.J, Willard, D.H, Wright, L.L, Zhou, H.Q. | | Deposit date: | 2005-02-10 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | P(2)-P(3) conformationally constrained ketoamide-based inhibitors of cathepsin K.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1POG
 
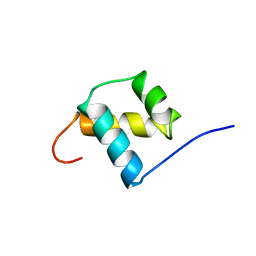 | | SOLUTION STRUCTURE OF THE OCT-1 POU-HOMEO DOMAIN DETERMINED BY NMR AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | OCT-1 POU HOMEODOMAIN DNA-BINDING PROTEIN | | Authors: | Cox, M, Van Tilborg, P.J.A, De Laat, W, Boelens, R, Van Leeuwen, H.C, Van Der Vliet, P.C, Kaptein, R. | | Deposit date: | 1994-10-12 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Oct-1 POU homeodomain determined by NMR and restrained molecular dynamics.
J.Biomol.NMR, 6, 1995
|
|
1H0W
 
 | | Human cyclin dependent protein kinase 2 in complex with the inhibitor 2-Amino-6-[cyclohex-3-enyl]methoxypurine | | Descriptor: | 1-AMINO-6-CYCLOHEX-3-ENYLMETHYLOXYPURINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Gibson, A.E, Arris, C.E, Bentley, J, Boyle, F.T, Curtin, N.J, Davies, T.G, Endicott, J.A, Golding, B.T, Grant, S, Griffin, R.J, Jewsbury, P, Johnson, L.N, Mesguiche, V, Newell, D.R, Noble, M.E.M, Tucker, J.A, Whitfield, H.J. | | Deposit date: | 2002-06-27 | | Release date: | 2003-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the ATP Ribose-Binding Domain of Cyclin-Dependent Kinases 1 and 2 with O(6)-Substituted Guanine Derivatives
J.Med.Chem., 45, 2002
|
|
