4DFA
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A/L36A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Clark, I.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4DGZ
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A/L125A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Nam, S.P, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
5BT5
 
 | | Crystal structure of BRD2 second bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Bromodomain-containing protein 2 | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of BRD2 second bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
4DO7
 
 | | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound zn, space group c2 | | Descriptor: | Amidohydrolase 2, SULFATE ION, ZINC ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-09 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound zn, space group c2
to be published
|
|
4DX7
 
 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | Acriflavine resistance protein B, DARPIN, DECANE, ... | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6FCU
 
 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with 4(NAG-NAMpentapeptide) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ACETATE ION, ALANINE, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4DF0
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Thermoproteus neutrophilus | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-22 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Thermoproteus neutrophilus
To be Published
|
|
5BRA
 
 | | Crystal Structure of a putative Periplasmic Solute binding protein (IPR025997) from Ochrobactrum Anthropi ATCC49188 (Oant_2843, TARGET EFI-511085) | | Descriptor: | Putative periplasmic binding protein with substrate ribose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.971 Å) | | Cite: | Crystal Structure of a putative Periplasmic Solute binding protein (IPR025997) from Ochrobactrum Anthropi ATCC49188(Oant_2843, TARGET EFI-511085)
To be published
|
|
5BT3
 
 | | Crystal structure of EP300 bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Histone acetyltransferase p300, ISOPROPYL ALCOHOL | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure of EP300 bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
5BV5
 
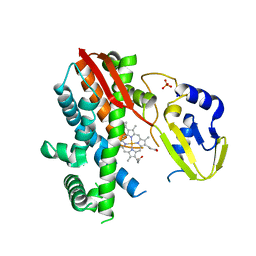 | | Structure of CYP119 with T213A and C317H mutations | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Cytochrome P450 119, PHOSPHATE ION, ... | | Authors: | Buller, A.R, Heel, T, McIntosh, J.A, Arnold, F.H. | | Deposit date: | 2015-06-04 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Adaptability Facilitates Histidine Heme Ligation in a Cytochrome P450.
J.Am.Chem.Soc., 137, 2015
|
|
4DGQ
 
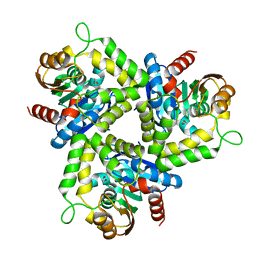 | | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Non-heme chloroperoxidase | | Authors: | Gardberg, A.S, Edwards, T.E, Abendroth, J.A, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-01-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia
To be Published
|
|
3C7H
 
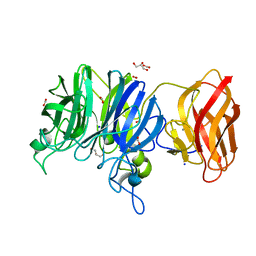 | | Crystal structure of glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with AXOS-4-0.5. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
4DF7
 
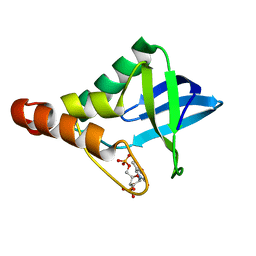 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23L/V99I at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4DFD
 
 | | Crystal structure of had family enzyme bt-2542 (target efi-501088) from bacteroides thetaiotaomicron, magnesium complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative haloacid dehalogenase-like hydrolase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
6F9G
 
 | | Ligand binding domain of P. putida KT2440 polyamine chemorecpetors McpU in complex putrescine. | | Descriptor: | 1,4-DIAMINOBUTANE, ACETATE ION, GLYCEROL, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M.T, Ortega, A, Martin-Mora, D, Corral-Lugo, A, Morel, B, Krell, T. | | Deposit date: | 2017-12-14 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Structural Basis for Polyamine Binding at the dCACHE Domain of the McpU Chemoreceptor from Pseudomonas putida.
J. Mol. Biol., 430, 2018
|
|
5G5K
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, BETA-HEXOSAMINIDASE | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-05-25 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5AOC
 
 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-valerate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
6F59
 
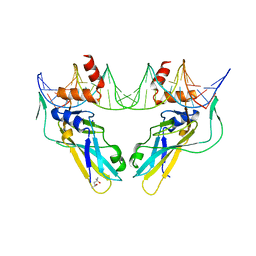 | | Crystal structure of human Brachyury (T) G177D variant in complex with DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Brachyury protein, DNA (26-MER), ... | | Authors: | Newman, J.A, Gavard, A.E, Krojer, T, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Crystal structure of human Brachyury (T) G177D variant in complex with DNA
To Be Published
|
|
3CRV
 
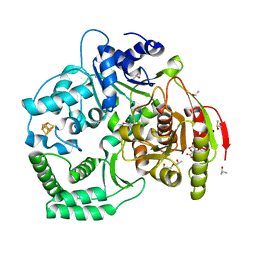 | | XPD_Helicase | | Descriptor: | CITRATE ANION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
5AND
 
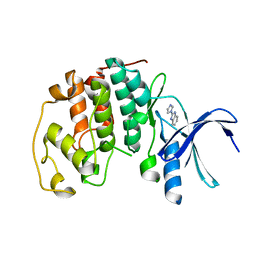 | | Crystal structure of CDK2 in complex with 2-imidazol-1-yl-1H- benzimidazole processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | 2-IMIDAZOL-1-YL-1H-BENZIMIDAZOLE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Zander, U, Hoffmann, G, Mathieu, M, Marquette, J.-P, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
4DX6
 
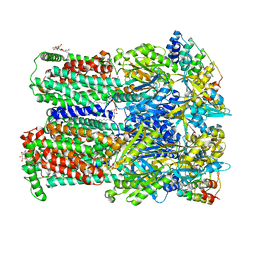 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | Acriflavine resistance protein B, DARPIN, DODECYL-BETA-D-MALTOSIDE | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6ESL
 
 | | Crystal structure of the Legionella pneumoppila LapA | | Descriptor: | Bacterial leucyl aminopeptidase, ZINC ION | | Authors: | Richardson, K, Garnett, J.A. | | Deposit date: | 2017-10-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Type II Secretion-Dependent Aminopeptidase LapA and Acyltransferase PlaC Are Redundant for Nutrient Acquisition duringLegionella pneumophilaIntracellular Infection of Amoebas.
MBio, 9, 2018
|
|
5AOA
 
 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-Propionate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
3CTW
 
 | |
5ANZ
 
 | |
