4IL2
 
 | | Crystal structure of D-mannonate dehydratase (rspA) from E. coli CFT073 (EFI TARGET EFI-501585) | | Descriptor: | MAGNESIUM ION, Starvation sensing protein rspA | | Authors: | Lukk, T, Wichelecki, D, Imker, H.J, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-12-28 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mannonate degradation pathway in E. coli CFT073
To be Published
|
|
5Q5G
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 117) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q5X
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 134) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4INF
 
 | | Crystal structure of amidohydrolase saro_0799 (target efi-505250) from novosphingobium aromaticivorans dsm 12444 with bound calcium | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of amidohydrolase sarp_0799 (target efi-505250) from novosphingobium aromaticivorans
To be Published
|
|
3P0W
 
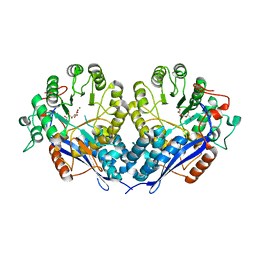 | | Crystal structure of D-Glucarate dehydratase from Ralstonia solanacearum complexed with Mg and D-glucarate | | Descriptor: | D-GLUCARATE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of D-Glucarate dehydratase from Ralstonia solanacearum complexed with Mg and D-glucarate
To be Published
|
|
3P3I
 
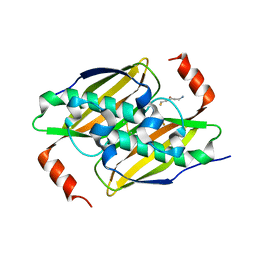 | | Crystal structure of the F36A mutant of the fluoroacetyl-CoA-specific thioesterase FlK in complex with fluoroacetate and CoA | | Descriptor: | COENZYME A, Fluoroacetyl coenzyme A thioesterase, fluoroacetic acid | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-04 | | Release date: | 2010-10-20 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
3P5Y
 
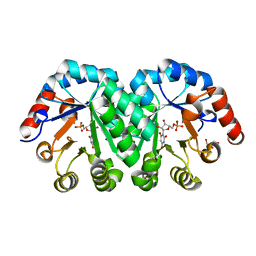 | | Crystal structure of the mutant T159A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
1QDD
 
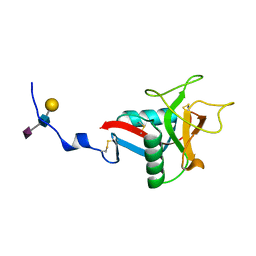 | | CRYSTAL STRUCTURE OF HUMAN LITHOSTATHINE TO 1.3 A RESOLUTION | | Descriptor: | LITHOSTATHINE, beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Gerbaud, V, Pignol, D, Loret, E, Bertrand, J.A, Berland, Y, Fontecilla-Camps, J.C, Canselier, J.P, Gabas, N, Verdier, J.M. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of calcite crystal growth inhibition by the N-terminal undecapeptide of lithostathine.
J.Biol.Chem., 275, 2000
|
|
1QGY
 
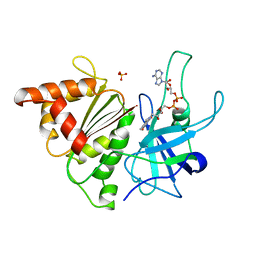 | | Ferredoxin:NADP+ reductase mutant with Lys 75 replaced by Glu (K75E) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP+ reductase, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of interactions for complex formation between Ferredoxin-NADP+ reductase and its protein partners
Proteins, 59, 2005
|
|
1QMP
 
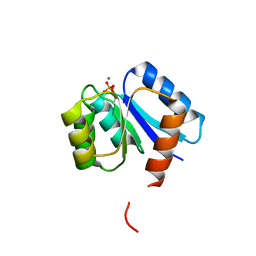 | | Phosphorylated aspartate in the crystal structure of the sporulation response regulator, Spo0A | | Descriptor: | CALCIUM ION, Stage 0 sporulation protein A | | Authors: | Lewis, R.J, Brannigan, J.A, Muchova, K, Barak, I, Wilkinson, A.J. | | Deposit date: | 1999-10-04 | | Release date: | 1999-11-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylated aspartate in the structure of a response regulator protein.
J. Mol. Biol., 294, 1999
|
|
5Q1J
 
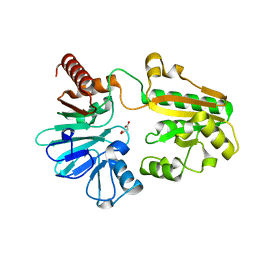 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMSOA000341b | | Descriptor: | DNA cross-link repair 1A protein, MALONATE ION, N-(2-hydroxyphenyl)acetamide, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1QMB
 
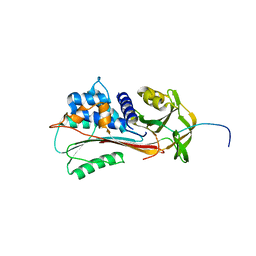 | | Cleaved alpha-1-antitrypsin polymer | | Descriptor: | ALPHA-1-ANTITRYPSIN | | Authors: | Huntington, J.A, Pannu, N.S, Hazes, B, Read, R.J, Lomas, D.A, Carrell, R.W. | | Deposit date: | 1999-09-24 | | Release date: | 2000-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A 2.6A Structure of a Serpin Polymer and Implications for Conformational Disease
J.Mol.Biol., 293, 1999
|
|
3OSO
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5Q1L
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000073a | | Descriptor: | (6~{R})-6-(4-methoxyphenyl)-2-oxidanylidene-5,6-dihydro-1~{H}-pyrimidine-4-carboxylic acid, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3OZY
 
 | | Crystal structure of enolase superfamily member from Bordetella bronchiseptica complexed with Mg and m-Xylarate | | Descriptor: | ACETIC ACID, D-xylaric acid, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-09-27 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2994 Å) | | Cite: | Crystal structure of enolase superfamily member from Bordetella bronchiseptica complexed with Mg and m-Xylarate
To be Published
|
|
4IKH
 
 | | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens pf-5, target efi-900003, with two glutathione bound | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-26 | | Release date: | 2013-01-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens pf-5, target efi-900003, with two glutathione bound
To be Published
|
|
5Q1Y
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000032a | | Descriptor: | 2-[cyclohexyl(methylsulfonyl)amino]ethanamide, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q2F
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 7) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q2U
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 22) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q3B
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 39) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q3Q
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 54) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4IGS
 
 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0064 | | Descriptor: | 2,2',3,3',5,5',6,6'-octafluorobiphenyl-4,4'-diol, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Porte, S, de Lera, A.R, Martin, M.J, de la Fuente, J.A, Klebe, G, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2012-12-18 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IDG
 
 | | Crystal structure of a short-chain dehydrogenase/reductase superfamily protein from agrobacterium tumefaciens (TARGET EFI-506441) with bound NAD, monoclinic form 2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-12 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a short-chain dehydrogenase/reductase superfamily protein from agrobacterium tumefaciens (TARGET EFI-506441) with bound NAD, monoclinic form 2
To be Published
|
|
3OSA
 
 | | Estrogen Receptor | | Descriptor: | 4-[1-(3-methylbut-2-en-1-yl)-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor | | Authors: | Bruning, J, Parent, A.A, Gil, G, Zhao, M, Nowak, J, Pace, M.C, Smith, C.L, Afonine, P.V, Adams, P.D, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Coupling of receptor conformation and ligand orientation determine graded activity.
Nat.Chem.Biol., 6, 2010
|
|
4II7
 
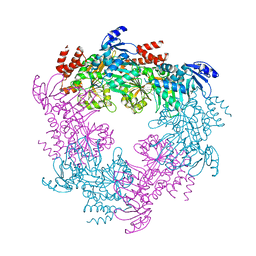 | | Archaellum Assembly ATPase FlaI | | Descriptor: | FlaI ATPase | | Authors: | Reindl, S, Williams, G.J, Tainer, J.A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Insights into FlaI Functions in Archaeal Motor Assembly and Motility from Structures, Conformations, and Genetics.
Mol.Cell, 49, 2013
|
|
