4K6D
 
 | | Crystal structure of Staphylococcal nuclease variant V23M/T62F at cryogenic temperature | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PHOSPHATE ION, ... | | 著者 | Caro, J.A, Flores, E, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | 登録日 | 2013-04-15 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Pressure effects on proteins
To be Published
|
|
4KQX
 
 | | Mutant Slackia exigua KARI DDV in complex with NAD and an inhibitor | | 分子名称: | HISTIDINE, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | 著者 | Brinkmann-Chen, S, Flock, T, Cahn, J.K.B, Snow, C.D, Brustad, E.M, Mcintosh, J.A, Meinhold, P, Zhang, L, Arnold, F.H. | | 登録日 | 2013-05-15 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KDX
 
 | | Crystal structure of a glutathione transferase family member from burkholderia graminis, target efi-507264, bound gsh, ordered domains, space group p21, form(1) | | 分子名称: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase domain | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-04-25 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Crystal structure of a glutathione transferase family member from burkholderia graminis, target efi-507264, bound gsh, ordered domains, space group P21, form(1)
To be Published
|
|
7M90
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 50 | | 分子名称: | 3C-like proteinase, 5-(3-{3-chloro-5-[2-(3-oxopiperazin-1-yl)ethoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | 著者 | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2021-03-30 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8Y
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 15 | | 分子名称: | 3C-like proteinase, 5-{3-[3-chloro-5-(2-phenylethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | 著者 | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2021-03-30 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8O
 
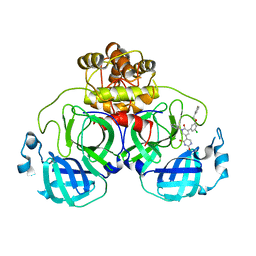 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 19 | | 分子名称: | 3C-like proteinase, 5-(3-{3-chloro-5-[(3-fluorophenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | 著者 | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2021-03-30 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M91
 
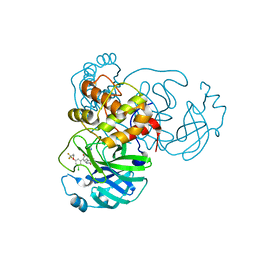 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 25 | | 分子名称: | 3C-like proteinase, 5-{3-[3-chloro-5-(3,3,3-trifluoropropoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | 著者 | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2021-03-30 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8X
 
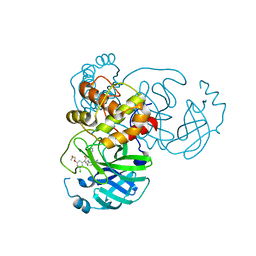 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 6 | | 分子名称: | 2-{3-[3-chloro-5-(2-methoxyethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}benzonitrile, 3C-like proteinase | | 著者 | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2021-03-30 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8Z
 
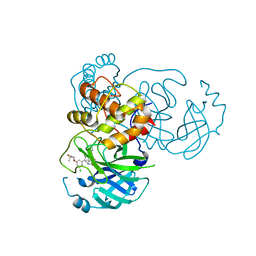 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 29 | | 分子名称: | 3C-like proteinase, 5-{3-[3-chloro-5-(3-hydroxy-3-methylbutoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | 著者 | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2021-03-30 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8P
 
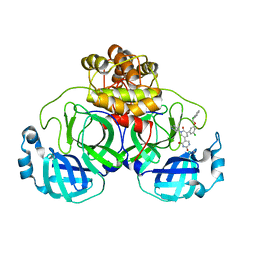 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 23 | | 分子名称: | 3C-like proteinase, 5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | 著者 | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2021-03-30 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8N
 
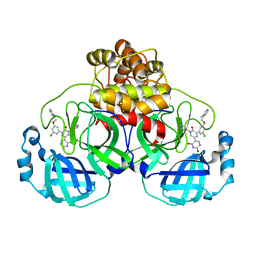 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 16 | | 分子名称: | 3C-like proteinase, 5-(3-{3-chloro-5-[(2-methylphenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | 著者 | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2021-03-30 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
2AYR
 
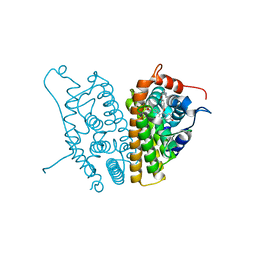 | | A SERM Designed for the Treatment of Uterine Leiomyoma with Unique Tissue Specificity for Uterus and Ovaries in Rats | | 分子名称: | 6-(4-METHYLSULFONYL-PHENYL)-5-[4-(2-PIPERIDIN-1-YLETHOXY)PHENOXY]NAPHTHALEN-2-OL, Estrogen receptor | | 著者 | Hummel, C.W, Geiser, A.G, Bryant, H.U, Cohen, I.R, Dally, R.D, Fong, K.C, Frank, S.A, Hinklin, R, Jones, S.A, Lewis, G, McCann, D.J, Shepherd, T.A, Tian, H, Rudman, D.G, Wallace, O.B, Wang, Y, Dodge, J.A. | | 登録日 | 2005-09-07 | | 公開日 | 2005-11-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A selective estrogen receptor modulator designed for the treatment of uterine leiomyoma with unique tissue specificity for uterus and ovaries in rats
J.Med.Chem., 48, 2005
|
|
1LGF
 
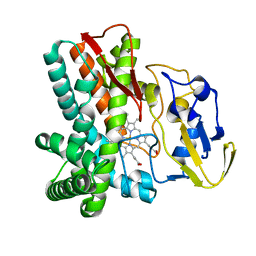 | | Crystal structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction During Vancomycin Biosynthesis | | 分子名称: | P450 monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Pylypenko, O, Zerbe, K, Vitali, F, Zhang, W, Vrijbloed, J.W, Robinson, J.A, Schlichting, I. | | 登録日 | 2002-04-15 | | 公開日 | 2002-12-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction during Vancomycin Biosynthesis.
J.Biol.Chem., 277, 2002
|
|
1KYN
 
 | | Cathepsin-G | | 分子名称: | (2-NAPHTHALEN-2-YL-1-NAPHTHALEN-1-YL-2-OXO-ETHYL)-PHOSPHONIC ACID, cathepsin G | | 著者 | Greco, M.N, Hawkins, M.J, Powell, E.T, Almond Jr, H.R, Corcoran, T.W, De Garavilla, L, Kauffman, J.A, Recacha, R, Chattopadhyay, D, Andrade-Gordon, P, Maryanoff, B.E. | | 登録日 | 2002-02-05 | | 公開日 | 2002-05-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Nonpeptide inhibitors of cathepsin G: optimization of a novel beta-ketophosphonic acid lead by structure-based drug design.
J.Am.Chem.Soc., 124, 2002
|
|
1YU4
 
 | | Major Tropism Determinant U1 Variant | | 分子名称: | MAGNESIUM ION, Major Tropism Determinant (Mtd-U1) | | 著者 | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | 登録日 | 2005-02-11 | | 公開日 | 2005-09-20 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YOK
 
 | | crystal structure of human LRH-1 bound with TIF-2 peptide and phosphatidylglycerol | | 分子名称: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Nuclear receptor coactivator 2, Orphan nuclear receptor NR5A2 | | 著者 | Krylova, I.N, Sablin, E.P, Moore, J, Xu, R.X, Waitt, G.M, MacKay, J.A, Juzumiene, D, Bynum, J.M, Madauss, K, Montana, V, Lebedeva, L, Suzawa, M, Williams, J.D, Williams, S.P, Guy, R.K, Thornton, J.W, Fletterick, R.J, Willson, T.M, Ingraham, H.A. | | 登録日 | 2005-01-27 | | 公開日 | 2005-07-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1.
Cell(Cambridge,Mass.), 120, 2005
|
|
4KC1
 
 | | Structure of the blood group glycosyltransferase AAglyB in complex with a pyridine inhibitor as a neutral pyrophosphate surrogate | | 分子名称: | 6-(1-beta-D-Glucopyranosyloxymethyl)-N-(5'-deoxyluridine-5'-yl)picolinamide, Fucosylglycoprotein alpha-N-acetylgalactosaminyltransferase, MANGANESE (II) ION, ... | | 著者 | Cuesta-Seijo, J.A, Wang, S, Lafont, D, Vidal, S, Palcic, M.M. | | 登録日 | 2013-04-24 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Design of glycosyltransferase inhibitors: pyridine as a pyrophosphate surrogate.
Chemistry, 19, 2013
|
|
1YQV
 
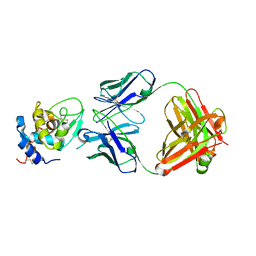 | | The crystal structure of the antibody Fab HyHEL5 complex with lysozyme at 1.7A resolution | | 分子名称: | Hen Egg White Lysozyme, HyHEL-5 Antibody Heavy Chain, HyHEL-5 Antibody Light Chain | | 著者 | Cohen, G.H, Silverton, E.W, Padlan, E.A, Dyda, F, Wibbenmeyer, J.A, Wilson, R.C, Davies, D.R. | | 登録日 | 2005-02-02 | | 公開日 | 2005-04-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Water molecules in the antibody-antigen interface of the structure of the Fab HyHEL-5-lysozyme complex at 1.7 A resolution: comparison with results from isothermal titration calorimetry.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4K5X
 
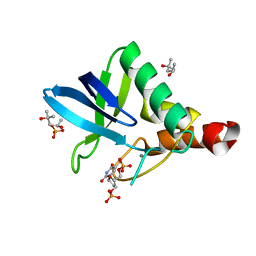 | | Crystal structure of Staphylococcal nuclease variant V23M/L36F at cryogenic temperature | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, PHOSPHATE ION, ... | | 著者 | Caro, J.A, Flores, E, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | 登録日 | 2013-04-15 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Pressure effects on proteins
To be Published
|
|
5AIG
 
 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. Tomsk-sample- Valpromide complex | | 分子名称: | 2-PROPYLPENTANAMIDE, DIMETHYL SULFOXIDE, LIMONENE-1,2-EPOXIDE HYDROLASE | | 著者 | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | 登録日 | 2015-02-13 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
5AII
 
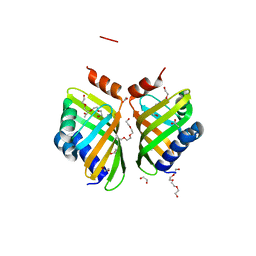 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. CH55-sample-PEG complex | | 分子名称: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | 著者 | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | 登録日 | 2015-02-13 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
4KDU
 
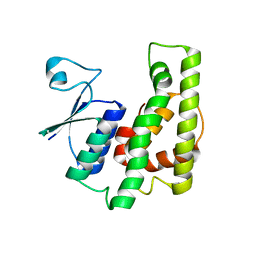 | | Crystal structure of a glutathione transferase family member from Burkholderia graminis, target efi-507264, no gsh, ordered domains, space group P21, form(1) | | 分子名称: | Glutathione S-transferase domain | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-04-25 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of a glutathione transferase family member from Burkholderia graminis, target efi-507264, no gsh, ordered domains, space group P21, form(1)
To be Published
|
|
5AHR
 
 | | Crystal structure of human DNA cross-link repair 1A, crystal form B | | 分子名称: | 1,2-ETHANEDIOL, DNA CROSS-LINK REPAIR 1A PROTEIN, ZINC ION | | 著者 | Allerston, C.K, Newman, J.A, Vollmar, M, Goubin, S, Forese, D.S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2015-02-06 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | The Structures of the Snm1A and Snm1B/Apollo Nuclease Domains Reveal a Potential Basis for Their Distinct DNA Processing Activities.
Nucleic Acids Res., 43, 2015
|
|
7P92
 
 | | TmHydABC- T. maritima bifurcating hydrogenase with bridge domain up | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | 著者 | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | 登録日 | 2021-07-23 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
1L8D
 
 | | Rad50 coiled-coil Zn hook | | 分子名称: | CITRIC ACID, DNA double-strand break repair rad50 ATPase, MERCURY (II) ION, ... | | 著者 | Hopfner, K.P, Tainer, J.A. | | 登録日 | 2002-03-20 | | 公開日 | 2002-08-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair.
Nature, 418, 2002
|
|
