5AG5
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | 分子名称: | (2R)-3-benzyl-2-(1H-indazol-5-yl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | 著者 | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | 登録日 | 2015-01-29 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
5AG6
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | 分子名称: | (2R)-2-(4-hydroxy-3-methoxyphenyl)-3-(pyridin-2-ylmethyl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | 著者 | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | 登録日 | 2015-01-29 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
1KWP
 
 | | Crystal Structure of MAPKAP2 | | 分子名称: | MAP Kinase Activated Protein Kinase 2, MERCURY (II) ION | | 著者 | Meng, W, Swenson, L.L, Fitzgibbon, M.J, Hayakawa, K, ter Haar, E, Behrens, A.E, Fulghum, J.R, Lippke, J.A. | | 登録日 | 2002-01-30 | | 公開日 | 2002-09-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of Mitogen-activated Protein Kinase-activated Protein (MAPKAP) Kinase 2 Suggests a Bifunctional Switch That
Couples Kinase Activation with Nuclear Export
J.Biol.Chem., 277, 2002
|
|
1XT7
 
 | | Daptomycin NMR Structure | | 分子名称: | DAPTOMYCIN, DECANOIC ACID | | 著者 | Ball, L.-J, Goult, C.M, Donarski, J.A, Micklefield, J, Ramesh, V. | | 登録日 | 2004-10-21 | | 公開日 | 2004-11-16 | | 最終更新日 | 2012-12-12 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure Determination and Calcium Binding Effects of Lipopeptide Antibiotic Daptomycin
Org.Biomol.Chem., 2, 2004
|
|
5CFL
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in complex with 3', 3' c-di-GMP, c[G(3', 5')pG(3', 5')p] | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CITRATE ANION, Stimulator of Interferon Genes | | 著者 | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | 登録日 | 2015-07-08 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.836 Å) | | 主引用文献 | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
1XVN
 
 | |
5CH7
 
 | | Crystal structure of the perchlorate reductase PcrAB - Phe164 gate switch intermediate - from Azospira suillum PS | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, ... | | 著者 | Tsai, C.-L, Tainer, J.A. | | 登録日 | 2015-07-10 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
5CM6
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOALTEROMONAS ATLANTICA T6c(Patl_2292, TARGET EFI-510180) WITH BOUND SODIUM AND PYRUVATE | | 分子名称: | PYRUVIC ACID, SODIUM ION, TRAP dicarboxylate transporter-DctP subunit | | 著者 | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2015-07-16 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOALTEROMONAS ATLANTICA T6c(Patl_2292, TARGET EFI-510180) WITH BOUND SODIUM AND PYRUVATE
To be published
|
|
1KLC
 
 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | TRANSFORMING GROWTH FACTOR-BETA 1 | | 著者 | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | 登録日 | 1996-01-16 | | 公開日 | 1996-08-17 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
5CNJ
 
 | | mGlur2 with glutamate analog | | 分子名称: | (1R,2S,4R,5R,6R)-2-amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | 著者 | Monn, J.A, Clawson, D.K. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
J.Med.Chem., 58, 2015
|
|
1KLD
 
 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 18-33 OF 33 STRUCTURES | | 分子名称: | TRANSFORMING GROWTH FACTOR-BETA 1 | | 著者 | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | 登録日 | 1996-01-16 | | 公開日 | 1996-08-17 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
4J9X
 
 | | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with trans-4-hydroxy-l-proline | | 分子名称: | 4-HYDROXYPROLINE, Proline racemase family protein, SODIUM ION | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-02-17 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with trans-4-hydroxy-l-proline
To be Published
|
|
7MGQ
 
 | | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans | | 分子名称: | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase, MAGNESIUM ION | | 著者 | Wizrah, M.S, Chua, S.M.H, Luo, Z, Manik, M.K, Pan, M, Whyte, J.M, Robertson, A.B, Kappler, U, Kobe, B, Fraser, J.A. | | 登録日 | 2021-04-13 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans.
J.Biol.Chem., 298, 2022
|
|
4JDP
 
 | | Crystal structure of probable p-nitrophenyl phosphatase (pho2) (target EFI-501307) from Archaeoglobus fulgidus DSM 4304 with Magnesium bound | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, p-nitrophenyl phosphatase (Pho2) | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Allen, K.N, Dunaway-Mariano, D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-02-25 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of probable p-nitrophenyl phosphatase from Archaeoglobus fulgidus.
To be Published
|
|
5CFR
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in apo 'unrotated' closed conformation | | 分子名称: | CALCIUM ION, Stimulator of Interferon Genes | | 著者 | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | 登録日 | 2015-07-08 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
4KEM
 
 | | Crystal structure of a tartrate dehydratase from azospirillum, target efi-502395, with bound mg and a putative acrylate ion, ordered active site | | 分子名称: | ACRYLIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Vetting, M.W, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-04-25 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal structure of a tartrate dehydratase from azospirillum, target efi-502395, with bound mg and a putative acrylate ion, ordered active site
To be Published
|
|
1XGL
 
 | | HUMAN INSULIN DISULFIDE ISOMER, NMR, 10 STRUCTURES | | 分子名称: | INSULIN | | 著者 | Hua, Q.X, Gozani, S.N, Chance, R.E, Hoffmann, J.A, Frank, B.H, Weiss, M.A. | | 登録日 | 1996-10-10 | | 公開日 | 1997-04-01 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a protein in a kinetic trap.
Nat.Struct.Biol., 2, 1995
|
|
5CGD
 
 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile - (HTL14242) | | 分子名称: | 3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, OLEIC ACID | | 著者 | Christopher, J.A, Aves, S.J, Bennett, K.A, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Okrasa, K, Serrano-Vega, M.J, Tehan, B.G, Wiggin, G.R, Congreve, M. | | 登録日 | 2015-07-09 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.603 Å) | | 主引用文献 | Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile).
J.Med.Chem., 58, 2015
|
|
5CGC
 
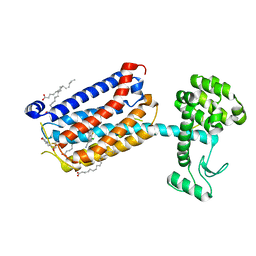 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | 著者 | Christopher, J.A, Aves, S.J, Bennett, K.A, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Okrasa, K, Serrano-Vega, M.J, Tehan, B.G, Wiggin, G.R, Congreve, M. | | 登録日 | 2015-07-09 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.101 Å) | | 主引用文献 | Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile).
J.Med.Chem., 58, 2015
|
|
5CHC
 
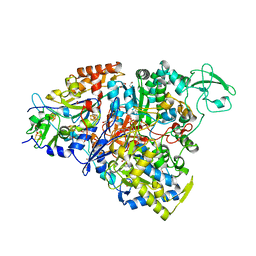 | | Crystal structure of the perchlorate reductase PcrAB - substrate analog SeO3 bound - from Azospira suillum PS | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BISELENITE ION, ... | | 著者 | Tsai, C.-L, Tainer, J.A. | | 登録日 | 2015-07-10 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
1MUN
 
 | |
5CNI
 
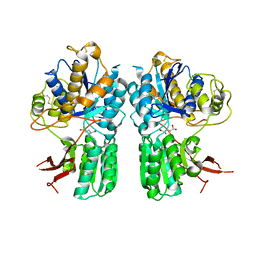 | | mGlu2 with Glutamate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | 著者 | Clawson, D.K, Atwell, S, Monn, J.A. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-02-15 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
J.Med.Chem., 58, 2015
|
|
1NAT
 
 | | CRYSTAL STRUCTURE OF SPOOF FROM BACILLUS SUBTILIS | | 分子名称: | SPORULATION RESPONSE REGULATORY PROTEIN | | 著者 | Madhusudan, Zapf, J, Hoch, J.A, Whiteley, J.M, Xuong, N.H, Varughese, K.I. | | 登録日 | 1997-09-09 | | 公開日 | 1998-10-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | A response regulatory protein with the site of phosphorylation blocked by an arginine interaction: crystal structure of Spo0F from Bacillus subtilis.
Biochemistry, 36, 1997
|
|
2BMV
 
 | | Apoflavodoxin from Helicobacter pylori | | 分子名称: | BENZAMIDINE, CHLORIDE ION, FLAVODOXIN | | 著者 | Martinez-Julvez, M, Hermoso, J.A, Sancho, J, Perez-Dorado, I, Cremades, N, Bueno, M. | | 登録日 | 2005-03-16 | | 公開日 | 2006-06-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Common Conformational Changes in Flavodoxins Induced by Fmn and Anion Binding: The Structure of Helicobacter Pylori Apoflavodoxin.
Proteins, 69, 2007
|
|
5C3F
 
 | | Crystal structure of Mcl-1 bound to BID-MM | | 分子名称: | BID-MM, GLYCEROL, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Miles, J.A, Yeo, D.J, Rowell, P, Rodriguez-Marin, S, Pask, C.M, Warriner, S.L, Edwards, T.A, Wilson, A.J. | | 登録日 | 2015-06-17 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Hydrocarbon constrained peptides - understanding preorganisation and binding affinity.
Chem Sci, 7, 2016
|
|
