2AFQ
 
 | |
2AF1
 
 | |
5UHW
 
 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+ and Magnesium | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase protein | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2017-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+ and Magnesium
To be published
|
|
7LJ1
 
 | | Human Prx1-Srx Decameric Complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Forshaw, T.E, Reisz, J.A, Nelson, K.J, Gumpena, R, Lawson, J.R, Jonsson, T, Wu, H, Clodfelter, J.E, Johnson, L, Furdui, C.M, Lowther, W.T. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Specificity of Human Sulfiredoxin for Reductant and Peroxiredoxin Oligomeric State.
Antioxidants (Basel), 10, 2021
|
|
258D
 
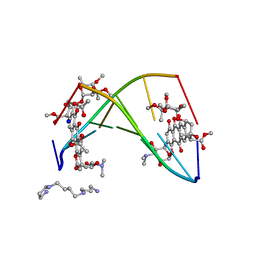 | | FACTORS AFFECTING SEQUENCE SELECTIVITY ON NOGALAMYCIN INTERCALATION: THE CRYSTAL STRUCTURE OF D(TGTACA)-NOGALAMYCIN | | Descriptor: | ACETATE ION, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3'), NOGALAMYCIN, ... | | Authors: | Smith, C.K, Brannigan, J.A, Moore, M.H. | | Deposit date: | 1996-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Factors affecting DNA sequence selectivity of nogalamycin intercalation: the crystal structure of d(TGTACA)2-nogalamycin2.
J.Mol.Biol., 263, 1996
|
|
7LWA
 
 | |
5CNI
 
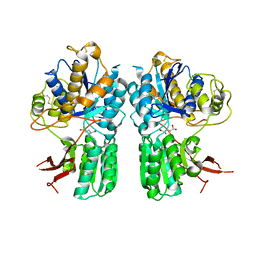 | | mGlu2 with Glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Clawson, D.K, Atwell, S, Monn, J.A. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
J.Med.Chem., 58, 2015
|
|
7LW7
 
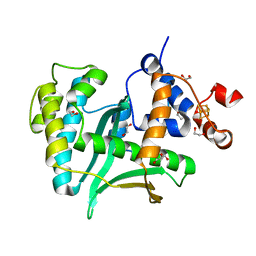 | | Human Exonuclease 5 crystal structure | | Descriptor: | 1,2-ETHANEDIOL, Exonuclease V, GLYCEROL, ... | | Authors: | Tsai, C.L, Tainer, J.A. | | Deposit date: | 2021-02-27 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart.
Mol.Cell, 81, 2021
|
|
4MGG
 
 | | Crystal structure of an enolase (mandelate racemase subgroup) from labrenzia aggregata iam 12614 (target nysgrc-012903) with bound mg, space group p212121 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Muconate lactonizing enzyme, ... | | Authors: | Vetting, M.W, Zhang, X, Wasserman, S.R, Morisco, L.L, Sojitra, S, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup) from labrenzia aggregata iam 12614 (target nysgrc-012903) with bound mg, space group p212121
To be Published
|
|
7LW9
 
 | | Human Exonuclease 5 crystal structure in complex with ssDNA, Sm, and Na | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*AP*TP*TP*GP*CP*TP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Tsai, C.L, Tainer, J.A. | | Deposit date: | 2021-02-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart.
Mol.Cell, 81, 2021
|
|
5UHZ
 
 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, D-Apionate and Magnesium | | Descriptor: | (3R,4R)-3,4-dihydroxy-4-(hydroxymethyl)oxolan-2-one, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2017-01-12 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, D-Apionate and Magnesium
To be published
|
|
4LTV
 
 | | Crystal structure of epi-isozizaene synthase from Streptomyces coelicolor A3(2) | | Descriptor: | Epi-isozizaene synthase, SULFATE ION | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
7LW8
 
 | | Human Exonuclease 5 crystal structure in complex with a ssDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Exonuclease V, ... | | Authors: | Tsai, C.L, Tainer, J.A. | | Deposit date: | 2021-02-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart.
Mol.Cell, 81, 2021
|
|
7LGI
 
 | | The haddock model of GDP KRas in complex with promazine using chemical shift perturbations and intermolecular NOEs | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, X, Gorfe, A.A, Putkey, J.A. | | Deposit date: | 2021-01-20 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Antipsychotic phenothiazine drugs bind to KRAS in vitro.
J.Biomol.Nmr, 75, 2021
|
|
2A8M
 
 | |
4LUI
 
 | | Crystal structure of Orotidine 5'-monophosphate decarboxylase from methanocaldococcus jannaschii | | Descriptor: | CHLORIDE ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-07-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Orotidine 5'-monophosphate decarboxylase from methanocaldococcus jannaschii
To be Published
|
|
2AB2
 
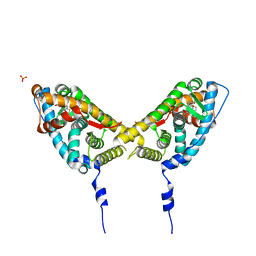 | | Mineralocorticoid Receptor Double Mutant with Bound Spironolactone | | Descriptor: | Mineralocorticoid receptor, SPIRONOLACTONE, SULFATE ION | | Authors: | Bledsoe, R.K, Madauss, K.P, Holt, J.A, Apolito, C.J, Lambert, M.H, Pearce, K.H, Stanley, T.B, Stewart, E.L, Trump, R.P, Willson, T.M, Williams, S.P. | | Deposit date: | 2005-07-14 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Ligand-mediated Hydrogen Bond Network Required for the Activation of the Mineralocorticoid Receptor
J.Biol.Chem., 280, 2005
|
|
2A8L
 
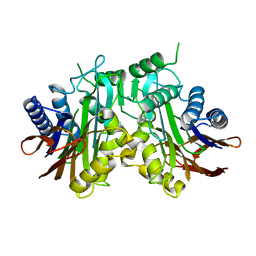 | |
4LZC
 
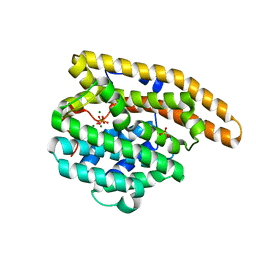 | | W325F Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate | | Descriptor: | Epi-isozizaene synthase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LUJ
 
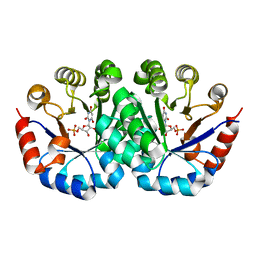 | | Crystal structure of Orotidine 5'-monophosphate decarboxylase from methanocaldococcus jannaschii complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-07-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Orotidine 5'-monophosphate decarboxylase from methanocaldococcus jannaschii complexed with inhibitor BMP
To be Published
|
|
4LV6
 
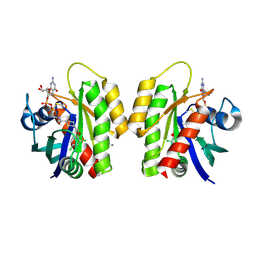 | | Crystal Structure of small molecule disulfide 4 covalently bound to K-Ras G12C | | Descriptor: | 1-[(2,4-dichlorophenoxy)acetyl]-N-(2-sulfanylethyl)piperidine-4-carboxamide, CALCIUM ION, GTPase KRas, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-26 | | Release date: | 2013-11-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
5UCE
 
 | | Solution NMR structure of the major species of DANCER-2, a dynamic and natively folded pentamutant of the B1 domain of streptococcal protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Damry, A.M, Davey, J.A, Goto, N.K, Chica, R.A. | | Deposit date: | 2016-12-22 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational design of proteins that exchange on functional timescales.
Nat. Chem. Biol., 13, 2017
|
|
5UB0
 
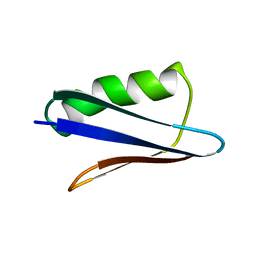 | | Solution NMR Structure of NERD-C, a natively folded tetramutant of the B1 domain of streptococcal protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Damry, A.M, Davey, J.A, Goto, N.K, Chica, R.A. | | Deposit date: | 2016-12-20 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational design of proteins that exchange on functional timescales.
Nat. Chem. Biol., 13, 2017
|
|
5UCF
 
 | | Solution NMR-derived model of the minor species of DANCER-2, a dynamic and natively folded pentamutant of the B1 domain of streptococcal protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Damry, A.M, Davey, J.A, Goto, N.K, Chica, R.A. | | Deposit date: | 2016-12-22 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational design of proteins that exchange on functional timescales.
Nat. Chem. Biol., 13, 2017
|
|
2B60
 
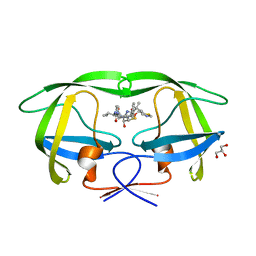 | | Structure of HIV-1 protease mutant bound to Ritonavir | | Descriptor: | GLYCEROL, Gag-Pol polyprotein, RITONAVIR | | Authors: | Clemente, J.C, Stow, L.R, Janka, L.K, Jeung, J.A, Desai, K.A, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2005-09-29 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In vivo, kinetic, and structural analysis of the development of ritonavir resistance
To be Published
|
|
