6VPS
 
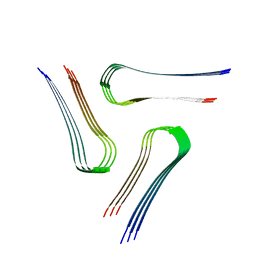 | | Cryo-EM structure of the amyloid core of Drosophila Orb2 isolated from head | | Descriptor: | Translational regulator orb2 | | Authors: | Hervas, R, Rau, M.J, Park, Y, Zhang, W, Murzin, A.G, Fitzpatrick, J.A.J, Scheres, S.H.W, Si, K. | | Deposit date: | 2020-02-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila
Science, 367, 2020
|
|
1FB7
 
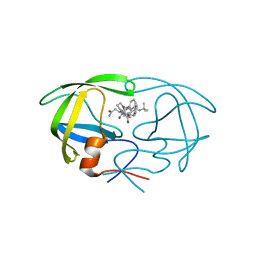 | | CRYSTAL STRUCTURE OF AN IN VIVO HIV-1 PROTEASE MUTANT IN COMPLEX WITH SAQUINAVIR: INSIGHTS INTO THE MECHANISMS OF DRUG RESISTANCE | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, HIV-1 PROTEASE | | Authors: | Hong, L, Zhang, X.C, Hartsuck, J.A, Tang, J. | | Deposit date: | 2000-07-14 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an in vivo HIV-1 protease mutant in complex with saquinavir: insights into the mechanisms of drug resistance.
Protein Sci., 9, 2000
|
|
1FBU
 
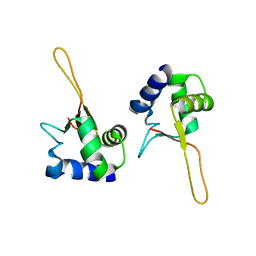 | | HEAT SHOCK TRANSCRIPTION FACTOR DNA BINDING DOMAIN | | Descriptor: | HEAT SHOCK FACTOR PROTEIN | | Authors: | Hardy, J.A, Nelson, H.C.M. | | Deposit date: | 2000-07-16 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proline in alpha-helical kink is required for folding kinetics but not for kinked structure, function, or stability of heat shock transcription factor.
Protein Sci., 9, 2000
|
|
4UPD
 
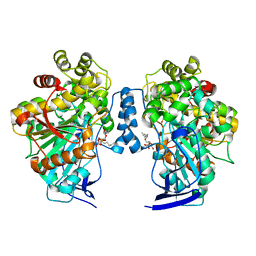 | | Open conformation of O. piceae sterol esterase mutant I544W | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, STEROL ESTERASE, TRIETHYLENE GLYCOL, ... | | Authors: | Gutierrez-Fernandez, J, Vaquero, M.E, Prieto, A, Barriuso, J, Gonzalez, M.J, Hermoso, J.A. | | Deposit date: | 2014-06-16 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Ophiostoma Piceae Sterol Esterase: Structural Insights Into Activation Mechanism and Product Release.
J.Struct.Biol., 187, 2014
|
|
4UHH
 
 | | Structural studies of a thermophilic esterase from Thermogutta terrifontis (cacodylate complex) | | Descriptor: | CACODYLIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Isupov, M.N, Bonch-Osmolovskaya, E, Littlechild, J.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structural Studies of a Thermophilic Esterase from a New Planctomycetes Species, Thermogutta Terrifontis.
FEBS J., 282, 2015
|
|
1FQR
 
 | | X-RAY CRYSTAL STRUCTURE OF COBALT-BOUND F93I/F95M/W97V CARBONIC ANHYDRASE (CAII) VARIANT | | Descriptor: | CARBONIC ANHYDRASE, COBALT (II) ION | | Authors: | Cox, J.D, Hunt, J.A, Compher, K.M, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2000-09-06 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural influence of hydrophobic core residues on metal binding and specificity in carbonic anhydrase II.
Biochemistry, 39, 2000
|
|
4UHP
 
 | | Crystal structure of the pyocin AP41 DNase-Immunity complex | | Descriptor: | BACTERIOCIN IMMUNITY PROTEIN, LARGE COMPONENT OF PYOCIN AP41 | | Authors: | Joshi, A, Chen, S, Wojdyla, J.A, Kaminska, R, Kleanthous, C. | | Deposit date: | 2015-03-25 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Ultra-High Affinity Protein-Protein Complexes of Pyocins S2 and Ap41 and Their Cognate Immunity Proteins from Pseudomonas Aeruginosa
J.Mol.Biol., 427, 2015
|
|
6THJ
 
 | | CBDP35 Native structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PlyP35 | | Authors: | Hermoso, J.A, Bartual, S.G. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CBDP35 Native structure
To Be Published
|
|
6T8Y
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
4UHU
 
 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase class A | | Descriptor: | ACETATE ION, FORMIC ACID, GNCA LACTAMASE W229D | | Authors: | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5LRB
 
 | | Plastidial phosphorylase from Barley in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-1,4 glucan phosphorylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cuesta-Seijo, J.A, Ruzanski, C, Kruzewicz, K, Palcic, M.M. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development.
PLoS ONE, 12, 2017
|
|
4UFW
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a pyridyl inhibitor (compound 22) | | Descriptor: | 2-oxopentadecyl-CoA, 4-chloranyl-N-[2-(3-methoxyphenyl)ethanimidoyl]-2-piperidin-4-yloxy-benzamide, CHLORIDE ION, ... | | Authors: | Yu, Z, Brannigan, J.A, Rangachari, K, Heal, W.P, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2015-03-19 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Pyridyl-Based Inhibitors of Plasmodium Falciparum N-Myristoyltransferase
Medchemcomm, 6, 2015
|
|
4UHE
 
 | | Structural studies of a thermophilic esterase from Thermogutta terrifontis (malate bound) | | Descriptor: | CHLORIDE ION, D-MALATE, ESTERASE, ... | | Authors: | Sayer, C, Isupov, M.N, Bonch-Osmolovskaya, E, Littlechild, J.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural Studies of a Thermophilic Esterase from a New Planctomycetes Species, Thermogutta Terrifontis.
FEBS J., 282, 2015
|
|
5M19
 
 | | Crystal structure of PBP2a from MRSA in the presence of Oxacillin ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
6TPN
 
 | | Crystal structure of the Orexin-2 receptor in complex with HTL6641 at 2.61 A resolution | | Descriptor: | 2-(5,6-dimethoxypyridin-3-yl)-1,1-bis(oxidanylidene)-4-[[2,4,6-tris(fluoranyl)phenyl]methyl]pyrido[2,3-e][1,2,4]thiadiazin-3-one, NITRATE ION, OLEIC ACID, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
4UHQ
 
 | | Crystal structure of the pyocin AP41 DNase | | Descriptor: | CITRIC ACID, LARGE COMPONENT OF PYOCIN AP41, NICKEL (II) ION | | Authors: | Joshi, A, Chen, S, Wojdyla, J.A, Kaminska, R, Kleanthous, C. | | Deposit date: | 2015-03-25 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of the Ultra-High Affinity Protein-Protein Complexes of Pyocins S2 and Ap41 and Their Cognate Immunity Proteins from Pseudomonas Aeruginosa
J.Mol.Biol., 427, 2015
|
|
4UHF
 
 | | Structural studies of a thermophilic esterase from Thermogutta terrifontis (L37A mutant with butyrate bound) | | Descriptor: | CACODYLIC ACID, ESTERASE, TRIETHYLENE GLYCOL, ... | | Authors: | Sayer, C, Isupov, M.N, Bonch-Osmolovskaya, E, Littlechild, J.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural Studies of a Thermophilic Esterase from a New Planctomycetes Species, Thermogutta Terrifontis.
FEBS J., 282, 2015
|
|
6TOD
 
 | | Crystal structure of the Orexin-1 receptor in complex with EMPA | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CITRIC ACID, N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
4UHD
 
 | | Structural studies of a thermophilic esterase from Thermogutta terrifontis (acetate bound) | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Isupov, M.N, Bonch-Osmolovskaya, E, Littlechild, J.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structural Studies of a Thermophilic Esterase from a New Planctomycetes Species, Thermogutta Terrifontis.
FEBS J., 282, 2015
|
|
6TQ9
 
 | | Crystal structure of the Orexin-1 receptor in complex with SB-408124 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 1-[6,8-bis(fluoranyl)-2-methyl-quinolin-4-yl]-3-[4-(dimethylamino)phenyl]urea, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
4UFX
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a pyridyl inhibitor (compound 19) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Yu, Z, Brannigan, J.A, Rangachari, K, Heal, W.P, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2015-03-19 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of Pyridyl-Based Inhibitors of Plasmodium Falciparum N-Myristoyltransferase
Medchemcomm, 6, 2015
|
|
6TOT
 
 | | Crystal structure of the Orexin-1 receptor in complex with lemborexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (1~{R},2~{S})-2-[(2,4-dimethylpyrimidin-5-yl)oxymethyl]-~{N}-(5-fluoranylpyridin-2-yl)-2-(3-fluorophenyl)cyclopropane-1-carboxamide, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
4UFV
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a pyridyl inhibitor (compound 18) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Yu, Z, Brannigan, J.A, Rangachari, K, Heal, W.P, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2015-03-19 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Pyridyl-Based Inhibitors of Plasmodium Falciparum N-Myristoyltransferase
Medchemcomm, 6, 2015
|
|
5M1A
 
 | | Crystal structure of PBP2a from MRSA in the presence of Ceftazidime ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
6TO7
 
 | | Crystal structure of the Orexin-1 receptor in complex with suvorexant at 2.29 A resolution | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CITRIC ACID, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
