1G00
 
 | |
3VKE
 
 | |
1GM8
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | CALCIUM ION, N-[(2S,4S,6R)-2-(DIHYDROXYMETHYL)-4-HYDROXY-3,3-DIMETHYL-7-OXO-4LAMBDA~4~-THIA-1-AZABICYCLO[3.2.0]HEPT-6-YL]-2-PHENYLAC ETAMIDE, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-11 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
6C3K
 
 | |
3V5C
 
 | | Crystal structure of the mutant E234A of Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Groninger-Poe, F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-12-16 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of the mutant E234A of Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg
To be Published
|
|
4E4J
 
 | | Crystal structure of arginine deiminase from Mycoplasma penetrans | | Descriptor: | Arginine deiminase, CHLORIDE ION | | Authors: | Benach, J, Gallego, P, Planell, R, Querol, E, Perez Pons, J.A, Reverter, D. | | Deposit date: | 2012-03-13 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma penetrans.
Plos One, 7, 2012
|
|
3UXW
 
 | | Crystal Structures of an A-T-hook/DNA complex | | Descriptor: | A-T hook peptide, dodecamer DNA | | Authors: | Fonfria-Subiros, E, Acosta-Reyes, F.J, Saperas, N, Pous, J, Subirana, J.A, Campos, J.L. | | Deposit date: | 2011-12-05 | | Release date: | 2012-05-23 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of a complex of DNA with one AT-hook of HMGA1.
Plos One, 7, 2012
|
|
1G3V
 
 | | CRYSTAL STRUCTURE OF NICKEL-D[CGTGTACACG]2 | | Descriptor: | 5'-D(*CP*GP*TP*GP*TP*AP*CP*AP*CP*G)-3', NICKEL (II) ION | | Authors: | Subirana, J.A, Abrescia, N.G.A. | | Deposit date: | 2000-10-25 | | Release date: | 2002-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nickel-guanine interactions in DNA: crystal structure of nickel-d[CGTGTACACG]2.
J.Biol.Inorg.Chem., 7, 2002
|
|
3UOT
 
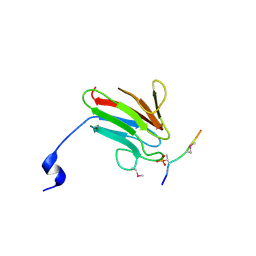 | | Crystal Structure of MDC1 FHA Domain in Complex with a Phosphorylated Peptide from the MDC1 N-terminus | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Clapperton, J.A, Lloyd, J, Haire, L.F, Li, J, Smerdon, S.J. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-28 | | Last modified: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator.
Nucleic Acids Res., 40, 2012
|
|
1G5S
 
 | | CRYSTAL STRUCTURE OF HUMAN CYCLIN DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH THE INHIBITOR H717 | | Descriptor: | 2-[TRANS-(4-AMINOCYCLOHEXYL)AMINO]-6-(BENZYL-AMINO)-9-CYCLOPENTYLPURINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Dreyer, M.K, Borcherding, D.R, Dumont, J.A, Peet, N.P, Tsay, J.T, Wright, P.S, Bitonti, A.J, Shen, J, Kim, S.-H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of human cyclin-dependent kinase 2 in complex with the adenine-derived inhibitor H717.
J.Med.Chem., 44, 2001
|
|
3VC6
 
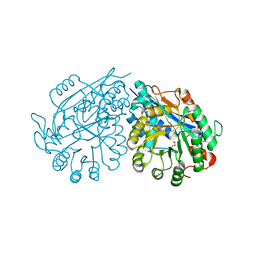 | | Crystal structure of enolase Tbis_1083(TARGET EFI-502310) FROM Thermobispora bispora DSM 43833 complexed with magnesium and formate | | Descriptor: | FORMIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of enolase Tbis_1083 FROM Thermobispora bispora DSM 43833
To be Published
|
|
1GKF
 
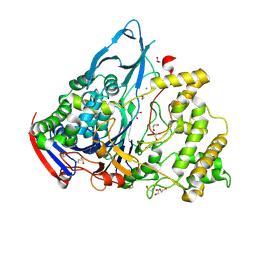 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-08-13 | | Release date: | 2002-01-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
3VC5
 
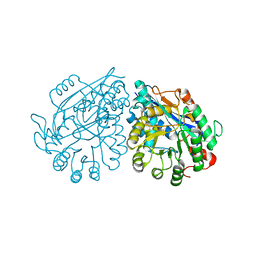 | | Crystal structure of enolase Tbis_1083(TARGET EFI-502310) FROM Thermobispora bispora DSM 43833 complexed with phosphate | | Descriptor: | Mandelate racemase/muconate lactonizing protein, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of enolase Tbis_1083 FROM Thermobispora bispora DSM 43833
To be Published
|
|
3H79
 
 | | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70 | | Descriptor: | THIOCYANATE ION, Thioredoxin-like protein | | Authors: | Santos, C.R, Fessel, M.R, Vieira, L.C, Krieger, M.A, Goldenberg, S, Guimaraes, B.G, Zanchin, N.I.T, Barbosa, J.A.R.G. | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70
TO BE PUBLISHED
|
|
3VDG
 
 | | Crystal structure of enolase MSMEG_6132 (TARGET EFI-502282) from Mycobacterium smegmatis str. MC2 155 complexed with formate and acetate | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of enolase MSMEG_6132 FROM Mycobacterium smegmatis
To be Published
|
|
1GK9
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-08-10 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme-Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
3VFH
 
 | |
1FZJ
 
 | | MHC CLASS I NATURAL MUTANT H-2KBM1 HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND VESICULAR STOMATITIS VIRUS NUCLEOPROTEIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rudolph, M.G, Speir, J.A, Brunmark, A, Mattsson, N, Jackson, M.R, Peterson, P.A, Teyton, L, Wilson, I.A. | | Deposit date: | 2000-10-03 | | Release date: | 2001-03-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of K(bm1) and K(bm8) reveal that subtle changes in the peptide environment impact thermostability and alloreactivity.
Immunity, 14, 2001
|
|
1IN7
 
 | | THERMOTOGA MARITIMA RUVB R170A | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1IN6
 
 | | THERMOTOGA MARITIMA RUVB K64R MUTANT | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
3TOU
 
 | | Crystal structure of GLUTATHIONE TRANSFERASE (TARGET EFI-501058) from Ralstonia solanacearum GMI1000 with GSH bound | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione s-transferase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of GLUTATHIONE S-TRANSFERASE from Ralstonia solanacearum
To be Published
|
|
1J8A
 
 | |
3TVU
 
 | | Crystal Structure of the humanized carboxyltransferase domain of yeast Acetyl-coA caroxylase in complex with compound 3 | | Descriptor: | 4-({4-[(2-methylquinolin-6-yl)methyl]piperidin-1-yl}carbonyl)-2-phenylquinoline, Acetyl-CoA carboxylase | | Authors: | Rajamohan, F, Marr, E, Reyes, A, Landro, J.A, Anderson, M.D, Corbett, J.W, Dirico, K.J, Harwood, J.H, Tu, M, Vajdos, F.F. | | Deposit date: | 2011-09-20 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-guided Inhibitor Design for Human Acetyl-coenzyme A Carboxylase by Interspecies Active Site Conversion.
J.Biol.Chem., 286, 2011
|
|
1IN5
 
 | | THERMOGOTA MARITIMA RUVB A156S MUTANT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Gonzalez, S, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
3TZ3
 
 | | Crystal Structure of the humanized carboxyltransferase domain of yeast Acetyl-coA caroxylase in complex with compound 2 | | Descriptor: | 6-{[1-(anthracen-9-ylcarbonyl)piperidin-4-yl]methyl}-2-methylquinoline, Acetyl-CoA carboxylase | | Authors: | Rajamohan, F, Marr, E, Reyes, A, Landro, J.A, Anderson, M.D, Corbett, J.W, Dirico, K.J, Harwood, J.H, Tu, M, Vajdos, F.F. | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided Inhibitor Design for Human Acetyl-coenzyme A Carboxylase by Interspecies Active Site Conversion.
J.Biol.Chem., 286, 2011
|
|
