4CYO
 
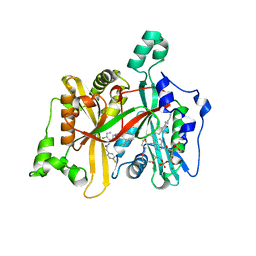 | | Leishmania major N-myristoyltransferase in complex with a hybrid inhibitor (compound 21). | | Descriptor: | GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, N-{5-[(3S,4R)-1-[(3R)-3-amino-4-(4-chlorophenyl)butanoyl]-4-(hydroxymethyl)pyrrolidin-3-yl]-2-chlorophenyl}-2-(4-fluorophenyl)acetamide, ... | | Authors: | Hutton, J.A, Goncalves, V, Brannigan, J.A, Paape, D, Waugh, T, Roberts, S.M, Bell, A.S, Wilkinson, A.J, Smith, D.F, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2014-04-14 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Design of Potent and Selective Leishmania N- Myristoyltransferase Inhibitors.
J.Med.Chem., 57, 2014
|
|
4CGP
 
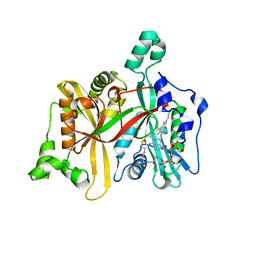 | | Leishmania major N-myristoyltransferase in complex with cofactor | | Descriptor: | GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, TETRADECANOYL-COA | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
4CGM
 
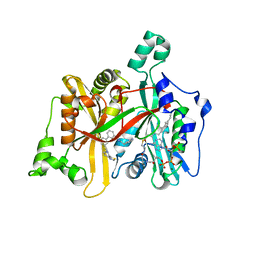 | | Leishmania major N-myristoyltransferase in complex with a biphenyl- derivative inhibitor | | Descriptor: | GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, N-[[3-[3-(6,7-dihydro-4H-[1,3]thiazolo[5,4-c]pyridin-5-ylmethyl)phenyl]phenyl]methyl]-2-pyridin-3-yl-ethanamine, ... | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
1MNS
 
 | |
4CGO
 
 | | Leishmania major N-myristoyltransferase in complex with a thienopyrimidine inhibitor | | Descriptor: | 3-[methyl-[2-[methyl-(1-methylpiperidin-4-yl)amino]thieno[3,2-d]pyrimidin-4-yl]amino]propanenitrile, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
1IKG
 
 | | MICHAELIS COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
6EK9
 
 | | Cytosolic copper storage protein Csp from Streptomyces lividans: Cu loaded form | | Descriptor: | COPPER (I) ION, Cytosolic copper storage protein | | Authors: | Straw, M.L, Chaplin, A.K, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-09-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans.
Metallomics, 10, 2018
|
|
5IXD
 
 | |
6EAX
 
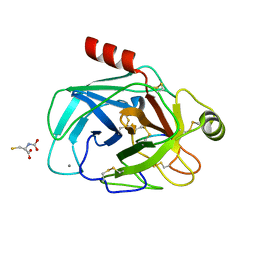 | | Crystallographic structure of the cyclic hexapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21 | | Descriptor: | CALCIUM ION, CYS-THR-LYS-SER-ILE-CYS, Cationic trypsin, ... | | Authors: | Fernandes, J.C, Valadares, N.F, Freitas, S.M, Barbosa, J.A.R.G. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.189 Å) | | Cite: | Crystallographic structure of the cyclic hexapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21
To Be Published
|
|
7RWN
 
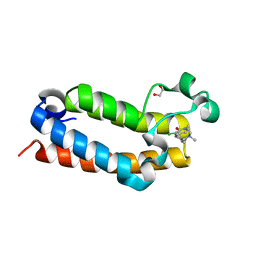 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
6EIY
 
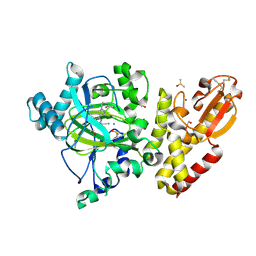 | | Crystal structure of KDM5B in complex with KDOPZ000034a. | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-~{N}-[2-[4-(3-cyano-7-oxidanylidene-6-propan-2-yl-4~{H}-pyrazolo[1,5-a]pyrimidin-5-yl)pyrazol-1-yl]ethyl]ethanamide, DIMETHYL SULFOXIDE, ... | | Authors: | Srikannathasan, V, Newman, J.A, Szykowska, A, Wright, M, Ruda, G.F, Vazquez-Rodriguez, S.A, Kupinska, K, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Huber, K, von Delft, F. | | Deposit date: | 2017-09-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of KDM5B in complex with KDOPZ000034a.
to be published
|
|
3NN8
 
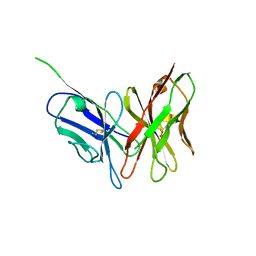 | |
4A2Z
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 4-METHOXY-2,3,6-TRIMETHYL-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
7RWP
 
 | | Crystal Structure of BPTF bromodomain in complex with 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one | | Descriptor: | 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWQ
 
 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWO
 
 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
6EJ0
 
 | | Crystal structure of KDM5B in complex with KDOPZ000049a. | | Descriptor: | 1,2-ETHANEDIOL, 7-oxidanylidene-6-propan-2-yl-5-[1-(1-prop-2-enoylpiperidin-4-yl)pyrazol-4-yl]-6~{H}-pyrazolo[1,5-a]pyrimidine-3-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Srikannathasan, V, Newman, J.A, Szykowska, A, Wright, M, Ruda, G.F, Vazquez-Rodriguez, S.A, Kupinska, K, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Huber, K, von Delft, F. | | Deposit date: | 2017-09-19 | | Release date: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of KDM5B in complex with KDOPZ000049a.
to be published
|
|
7ZMO
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-052 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-052, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZL2
 
 | | Crystal Structure of human Brachyury G177D variant in complex with Molpolrt-039-246-810 | | Descriptor: | T-box transcription factor T, UNKNOWN LIGAND | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of human Brachyury G177D variant in complex with Molpolrt-039-246-810
To Be Published
|
|
7ZKF
 
 | | Crystal Structure of human Brachyury G177D variant in complex with CF-5-86 | | Descriptor: | PHOSPHATE ION, T-box transcription factor T, UNKNOWN LIGAND | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of human Brachyury G177D variant in complex with CF-5-86
To Be Published
|
|
7ZMV
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G5-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMT
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G5-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZK2
 
 | | Crystal Structure of human Brachyury G177D variant in complex with CSC027898502 | | Descriptor: | N-[4-(2-morpholin-4-yl-1,3-thiazol-4-yl)phenyl]ethanamide, PHOSPHATE ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of human Brachyury G177D variant in complex with CSC027898502
To Be Published
|
|
7ZMP
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-055 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-055, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.626 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMR
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G2*-011 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G2*-011, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
