7Z49
 
 | | The capsid of bacteriophage SU10. | | Descriptor: | Major head protein | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-10-05 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
5A3F
 
 | | Crystal structure of the dynamin tetramer | | Descriptor: | DYNAMIN 3 | | Authors: | Reubold, T.F, Faelber, K, Plattner, N, Posor, Y, Branz, K, Curth, U, Schlegel, J, Anand, R, Manstein, D.J, Noe, F, Haucke, V, Daumke, O, Eschenburg, S. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the Dynamin Tetramer
Nature, 525, 2015
|
|
5K1G
 
 | | Crystal structure of reduced Prx3 from Vibrio vulnificus | | Descriptor: | 1-Cys peroxiredoxin | | Authors: | Ahn, J, Ha, N.-C. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Crystal structure of peroxiredoxin 3 fromVibrio vulnificusand its implications for scavenging peroxides and nitric oxide.
IUCrJ, 5, 2018
|
|
2ZV7
 
 | | Lyn Tyrosine Kinase Domain, apo form | | Descriptor: | Tyrosine-protein kinase Lyn | | Authors: | Williams, N.K, Rossjohn, J. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the Lyn Protein Tyrosine Kinase Domain in Its Apo- and Inhibitor-bound State
J.Biol.Chem., 284, 2009
|
|
7YRT
 
 | |
2ZJ9
 
 | | X-ray crystal structure of AmpC beta-Lactamase (AmpC(D)) from an Escherichia coli with a Tripeptide Deletion (Gly286 Ser287 Asp288) on the H10 Helix | | Descriptor: | AmpC, ISOPROPYL ALCOHOL, SODIUM ION | | Authors: | Yamaguchi, Y, Sato, G, Yamagata, Y, Wachino, J, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2008-02-29 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of AmpC beta-lactamase (AmpCD) from an Escherichia coli clinical isolate with a tripeptide deletion (Gly286-Ser287-Asp288) in the H10 helix
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5KBK
 
 | | Candida Albicans Superoxide Dismutase 5 (SOD5), E110A Mutant | | Descriptor: | COPPER (I) ION, Cell surface Cu-only superoxide dismutase 5, SULFATE ION | | Authors: | Galaleldeen, A, Peterson, R.L, Villarreal, J, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.411 Å) | | Cite: | The Phylogeny and Active Site Design of Eukaryotic Copper-only Superoxide Dismutases.
J.Biol.Chem., 291, 2016
|
|
5KBU
 
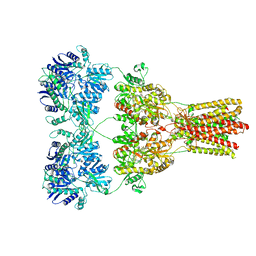 | | Cryo-EM structure of GluA2-2xSTZ complex at 7.8 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
3KR3
 
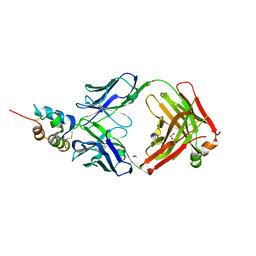 | | Crystal structure of IGF-II antibody complex | | Descriptor: | 1,2-ETHANEDIOL, Insulin-like growth factor II, antibody-Fab (heavy chain), ... | | Authors: | Peat, T.S, Newman, J, Adams, T.E. | | Deposit date: | 2009-11-17 | | Release date: | 2010-06-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A human monoclonal antibody against insulin-like growth factor-II blocks the growth of human hepatocellular carcinoma cell lines in vitro and in vivo.
Mol.Cancer Ther., 9, 2010
|
|
5DCA
 
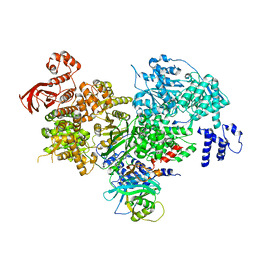 | | Crystal structure of yeast full length Brr2 in complex with Prp8 Jab1 domain | | Descriptor: | Pre-mRNA-splicing factor 8, Pre-mRNA-splicing helicase BRR2 | | Authors: | Absmeier, E, Wollenhaupt, J, Santos, K.F, Wahl, M.C. | | Deposit date: | 2015-08-23 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation.
Genes Dev., 29, 2015
|
|
1FWT
 
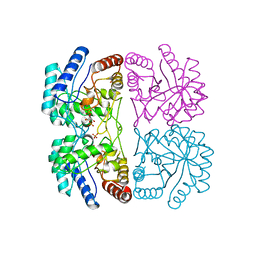 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP, E4P AND CADMIUM | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, CADMIUM ION, ERYTHOSE-4-PHOSPHATE, ... | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-24 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate and metal complexes of 3-deoxy-D-manno-octulosonate-8-phosphate synthase from Aquifex aeolicus at 1.9-A resolution. Implications for the condensation mechanism.
J.Biol.Chem., 276, 2001
|
|
7YQC
 
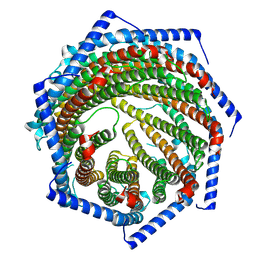 | | EM structure of human PA28gamma | | Descriptor: | Proteasome activator complex subunit 3 | | Authors: | Chen, D.-D, Hao, J, Shen, C.-H, Yun, C.-H. | | Deposit date: | 2022-08-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Atomic resolution Cryo-EM structure of human proteasome activator PA28 gamma.
Int.J.Biol.Macromol., 219, 2022
|
|
5KEP
 
 | | High resolution cryo-EM maps of Human Papillomavirus 16 reveal L2 location and heparin-induced conformational changes | | Descriptor: | Major capsid protein L1 | | Authors: | Guan, J, Bywaters, S.M, Brendle, S.A, Ashley, R.E, Makhov, A.M, Conway, J.F, Christensen, N.D, Hafenstein, S. | | Deposit date: | 2016-06-10 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryoelectron Microscopy Maps of Human Papillomavirus 16 Reveal L2 Densities and Heparin Binding Site.
Structure, 25, 2017
|
|
1SPQ
 
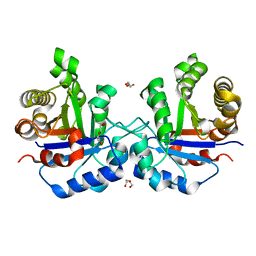 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | DI(HYDROXYETHYL)ETHER, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
8OK7
 
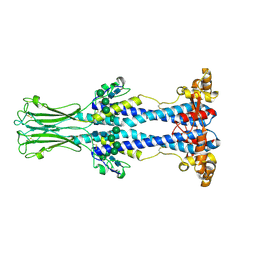 | | Variant Surface Glycoprotein VSG558 NTD | | Descriptor: | Variant surface glycoprotein 558, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zeelen, J.P, Stebbins, C.E, van Straaten, M, Zhong, J. | | Deposit date: | 2023-03-27 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
5CRB
 
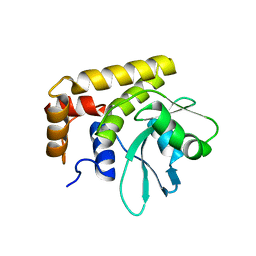 | | Crystal Structure of SdeA DUB | | Descriptor: | SdeA | | Authors: | Sheedlo, M.J, Qiu, J, Luo, Z.Q, Das, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5K7Y
 
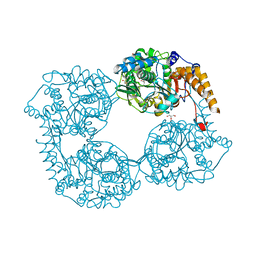 | | Crystal structure of enzyme in purine metabolism | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL | | Authors: | Skerlova, J, Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2016-05-27 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Oligomeric interface modulation causes misregulation of purine 5 -nucleotidase in relapsed leukemia.
Bmc Biol., 14, 2016
|
|
4QQ4
 
 | | CW-type zinc finger of MORC3 in complex with the amino terminus of histone H3 | | Descriptor: | CHLORIDE ION, Histone H3.3, MORC family CW-type zinc finger protein 3, ... | | Authors: | Liu, Y, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Family-wide Characterization of Histone Binding Abilities of Human CW Domain-containing Proteins.
J.Biol.Chem., 291, 2016
|
|
5CS4
 
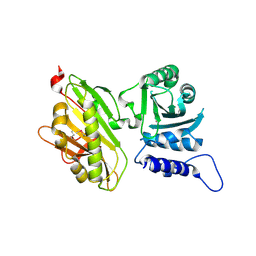 | |
1I5U
 
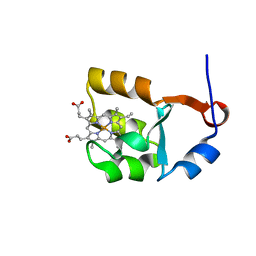 | | SOLUTION STRUCTURE OF CYTOCHROME B5 TRIPLE MUTANT (E48A/E56A/D60A) | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Qian, C, Yao, Y, Tang, W, Wang, J, Zhongxian, H. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of charged amino-acid mutation on the solution structure of cytochrome b(5) and binding between cytochrome b(5) and cytochrome c.
Protein Sci., 10, 2001
|
|
8OIE
 
 | | Iron Nitrogenase Complex from Rhodobacter capsulatus | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Schmidt, F.V, Schulz, L, Zarzycki, J, Prinz, S, Erb, T.J, Rebelein, J.G. | | Deposit date: | 2023-03-22 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural insights into the iron nitrogenase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1SH0
 
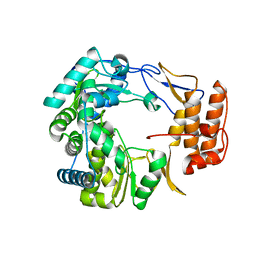 | | Crystal Structure of Norwalk Virus Polymerase (Triclinic) | | Descriptor: | RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
5CUQ
 
 | | Identification and characterization of novel broad spectrum inhibitors of the flavivirus methyltransferase | | Descriptor: | N,N'-BIS(4-AMINO-2-METHYLQUINOLIN-6-YL)UREA, Nonstructural protein NS5 | | Authors: | Brecher, B, Chen, H, Li, Z, Banavali, N.K, Jones, S.A, Zhang, J, Kramer, L.D, Li, H.M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Identification and Characterization of Novel Broad-Spectrum Inhibitors of the Flavivirus Methyltransferase.
Acs Infect Dis., 1, 2015
|
|
4QRS
 
 | | Crystal Structure of HLA B*0801 in complex with ELK_IYM, ELKRKMIYM | | Descriptor: | ACETATE ION, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Twist, K.-A, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-12-10 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular imprint of exposure to naturally occurring genetic variants of human cytomegalovirus on the T cell repertoire.
Sci Rep, 4, 2014
|
|
8JZZ
 
 | | Structure of human C5a-desArg bound human C5aR1 in complex with Go | | Descriptor: | Antibody fragment ScFv16, C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-07-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
