6FRF
 
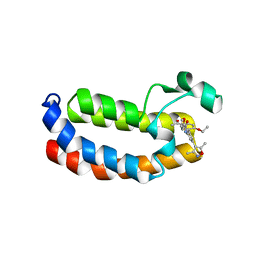 | | Crystal structure of CREBBP bromodomain complexd with PA10 | | Descriptor: | CREB-binding protein, ~{N}-[3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-(5-ethanoyl-2-ethoxy-phenyl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-15 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
7ZHH
 
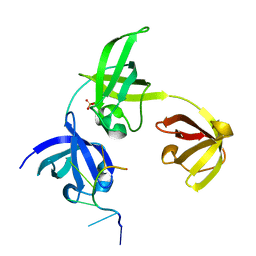 | | Complex structure of drosophila Unr CSD789 and a poly(A) RNA sequence | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, Upstream of N-ras, ... | | Authors: | Hollmann, N.M, Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res., 51, 2023
|
|
1AFV
 
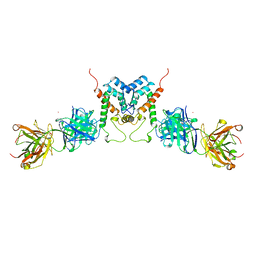 | | HIV-1 CAPSID PROTEIN (P24) COMPLEX WITH FAB25.3 | | Descriptor: | ANTIBODY FAB25.3 FRAGMENT (HEAVY CHAIN), ANTIBODY FAB25.3 FRAGMENT (LIGHT CHAIN), HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 CAPSID PROTEIN, ... | | Authors: | Momany, C, Kovari, L.C, Prongay, A.J, Keller, W, Gitti, R.K, Lee, B.M, Gorbalenya, A.E, Tong, L, Mcclure, J, Ehrlich, L.S, Summers, M.F, Carter, C, Rossmann, M.G. | | Deposit date: | 1997-03-14 | | Release date: | 1997-08-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of dimeric HIV-1 capsid protein.
Nat.Struct.Biol., 3, 1996
|
|
6TBB
 
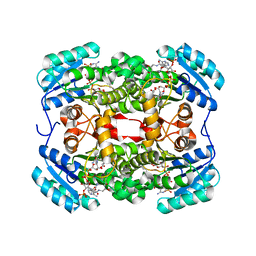 | | Crystal structure of S. aureus FabI in complex with NADPH and kalimantacin A (batumin) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], Kalimantacin, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Fage, C.D, Masschelein, J. | | Deposit date: | 2019-11-01 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Kalimantacin Polyketide Antibiotics Inhibit Fatty Acid Biosynthesis in Staphylococcus aureus by Targeting the Enoyl-Acyl Carrier Protein Binding Site of FabI.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6CNV
 
 | | INFLUENZA B/BRISBANE HEMAGGLUTININ FAB CR9115 SD84H COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR9114 Fab heavy chain, ... | | Authors: | Luo, J, Obmolova, G. | | Deposit date: | 2018-03-09 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Science, 362, 2018
|
|
5YJO
 
 | |
8U8M
 
 | | X-ray crystal structure of TEBP-1 MCD2 homodimer | | Descriptor: | COBALT (II) ION, Double-strand telomeric DNA-binding proteins 1 | | Authors: | Nandakumar, J, Padmanaban, S. | | Deposit date: | 2023-09-18 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Caenorhabditis elegans telomere-binding proteins TEBP-1 and TEBP-2 adapt the Myb module to dimerize and bind telomeric DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8I9Q
 
 | | The focused refinement of CCT4-PhLP2A from TRiC-PhLP2A complex in the open state | | Descriptor: | Phosducin-like protein 3, T-complex protein 1 subunit delta | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
6TEI
 
 | | Crystal structure of human protein kinase CK2alpha (CSNK2A1 gene product) in complex with the 2-aminothiazole-type inhibitor 17 | | Descriptor: | 3-[(4-pyridin-2-yl-1,3-thiazol-2-yl)amino]benzoic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Niefind, K, Lindenblatt, D, Jose, J, Applegate, V.M, Nickelsen, A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Structural and Mechanistic Basis of the Inhibitory Potency of Selected 2-Aminothiazole Compounds on Protein Kinase CK2.
J.Med.Chem., 63, 2020
|
|
7BHD
 
 | | FimH in complex with alpha1,6 core-fucosylated oligomannose-3, crystallized in the trigonal space group | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bridot, C, Bouckaert, J, Krammer, E.-M. | | Deposit date: | 2021-01-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
8IB8
 
 | | Human TRiC-PhLP2A-actin complex in the closed state | | Descriptor: | ACTB protein (Fragment), Phosducin-like protein 3, T-complex protein 1 subunit alpha, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-09 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
148D
 
 | |
1A95
 
 | | XPRTASE FROM E. COLI COMPLEXED WITH MG:CPRPP AND GUANINE | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, BORIC ACID, GUANINE, ... | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
6GB9
 
 | |
1IDK
 
 | | PECTIN LYASE A | | Descriptor: | PECTIN LYASE A | | Authors: | Mayans, O, Scott, M, Connerton, I, Gravesen, T, Benen, J, Visser, J, Pickersgill, R, Jenkins, J. | | Deposit date: | 1996-10-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases.
Structure, 5, 1997
|
|
6T5F
 
 | | Human 14-3-3 sigma fused to the StARD1 peptide including phosphoserine-195 | | Descriptor: | 14-3-3 protein sigma, StARD1 peptide | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Titterington, J, Antson, A.A. | | Deposit date: | 2019-10-16 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Molecular basis for the recognition of steroidogenic acute regulatory protein by the 14-3-3 protein family.
Febs J., 287, 2020
|
|
8TGN
 
 | | VMAT1 dimer with serotonin and reserpine | | Descriptor: | Chromaffin granule amine transporter, SEROTONIN, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
6T6H
 
 | | Apo structure of the Bottromycin epimerase BotH | | Descriptor: | BotH, SODIUM ION, SULFATE ION | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2019-10-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | The bottromycin epimerase BotH defines a group of atypical alpha / beta-hydrolase-fold enzymes.
Nat.Chem.Biol., 16, 2020
|
|
7C4U
 
 | | MicroED structure of orthorhombic Vancomycin at 1.2 A resolution | | Descriptor: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | Authors: | Fan, Q, Zhou, H, Li, X, Wang, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-08-12 | | Last modified: | 2021-03-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6CYD
 
 | | PDE2 in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 3-(hydroxymethyl)-6-methyl-1-{(1S)-1-[4-(trifluoromethyl)phenyl]ethyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, MAGNESIUM ION, ... | | Authors: | Lu, J. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-Guided Design and Procognitive Assessment of a Potent and Selective Phosphodiesterase 2A Inhibitor.
ACS Med Chem Lett, 9, 2018
|
|
6T6Z
 
 | |
8PP7
 
 | | human RYBP-PRC1 bound to mononucleosome | | Descriptor: | DNA (215-mer), E3 ubiquitin-protein ligase RING2, Histone H2A, ... | | Authors: | Ciapponi, M, Benda, C, Mueller, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis of the histone ubiquitination read-write mechanism of RYBP-PRC1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6MQW
 
 | |
8TQ7
 
 | | Crystal structure of Fab.34.2.12 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab 34.2.12 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
